This is probably the best way to spot bacteria in a fluid
Researchers at Stanford University have proposed a better, faster, and cheaper technique to detect bacteria in fluid samples. The new technique is not only limited to finding bacteria but also reveals the type of bacteria a blood or water sample has. According to the researchers, whereas conventional bacteria screening tests can take hours or even months (depending on the sample and type of bacteria), the proposed method can deliver results almost immediately.


Many of us don’t realize it but fast bacteria detection and identification is crucial for food safety, medical diagnosis, drug development, and various other purposes. For instance, quick bacteria detection methods can help us rapidly purify drinking water by spotting and killing harmful bacteria quickly. But as important as it is, detecting bacteria isn’t always trivial. While highlighting a side effect of conventional bacteria detection methods which are slow, the researchers said:
“Though in vitro liquid culturing is typically used for pathogen detection, it is estimated that less than 2% of all bacteria can be readily cultured using current laboratory protocols. Further, among that 2%, culturing can take hours to days depending on the bacterial species. In the case of diagnostics, broad-spectrum antibiotics are often administered while waiting for culture results, leading to an alarming rise in antibiotic-resistant bacteria.”
What’s more surprising is that the proposed method has been developed by combining three unrelated technologies; artificial intelligence, nanotechnology, and inkjet printing.
The science behind the new bacteria identification method
To study microscopic objects such as body cells or bacteria, scientists employ a technique called Raman spectroscopy that basically involves throwing light at objects and examining the reflected light rays. So for example, when a scientist shines a laser light on a red blood cell and bacteria, the light reflected by the former would be different from that reflected by the latter because they have different spectral fingerprints.

This is how a scientist is able to differentiate between different microscopic things in our bodies. However, when the same technique is used to identify bacteria in a blood droplet, things get complicated because even a drop of blood contains billions of cells, with each having its own distinct spectral fingerprint. First author and a Ph.D. candidate at Stanford University, Fareeha Safir said:
“Not only does each type of bacterium demonstrate unique patterns of light but virtually every other molecule or cell in a given sample does too. Red blood cells, white blood cells, and other components in the sample are sending back their own signals, making it hard if not impossible to distinguish the microbial patterns from the noise of other cells.”
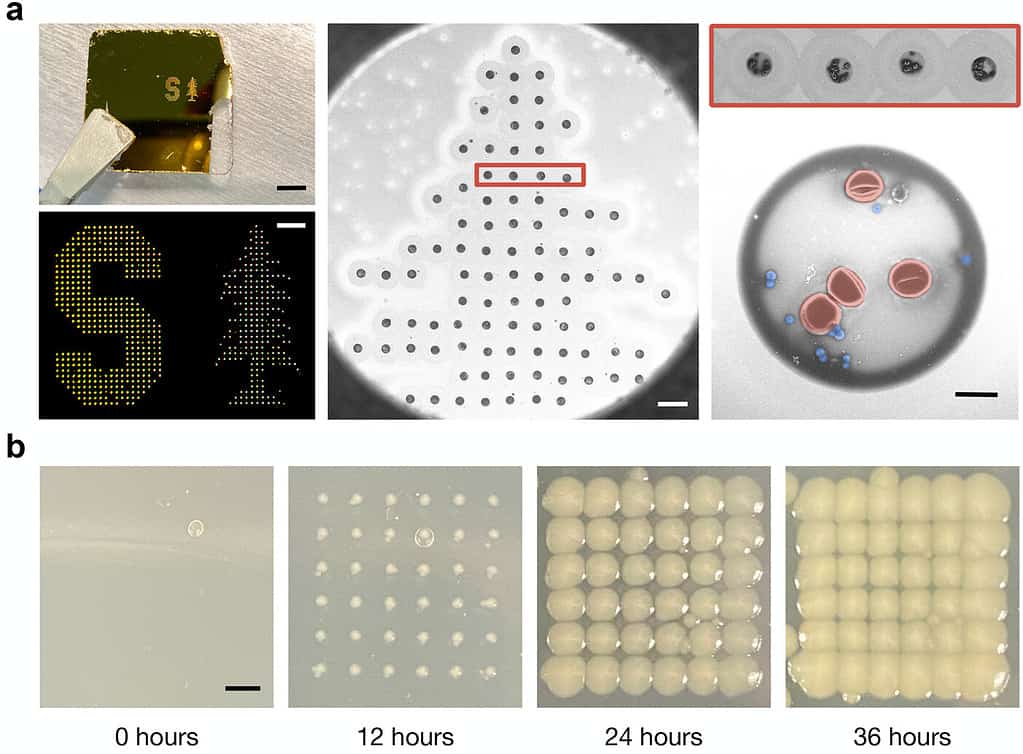
Usually, to amplify the bacteria signal out of all other signals, scientists culture bacteria in a lab and this method ends up taking a lot of time. The new method does not involve any in vitro bacteria culturing, instead, it focuses on decreasing the sample size to the extent that it becomes easy to spot the bacteria. They used a modified inkjet printing technology to print numerous tiny blood dots from the tiny blood droplet (having a size of about a millimeter).
Each of these dots was about a billion times smaller than the droplet and contained only a few dozen cells. Next, they inserted nanorods made of gold into the droplets. When laser light was shot at the dots, these nanorods directed the laser light toward the bacteria and amplified the spectral signal of the microorganisms by 1500 times.
The researchers then employed an AI program to study the different spectra reflected by the dots and successfully identified the different types of bacteria that were present in the blood drop sample.
“We show that we generate these consistent Raman spectra from Gram-positive and Gram-negative bacteria as well as from RBCs and can differentiate spectra. Finally, we demonstrate that we can identify distinct cell types present in droplets printed from a mixture of cell lines using machine-learning algorithms,” the researchers note.
The significance of AI-assisted Raman spectroscopy
The study authors believe that their research could advance Raman spectroscopy-based clinical diagnosis, research, and disease management practices. This approach represents a reliable, accurate, and fast way to detect harmful bacteria in any fluid sample ranging from water to blood and mucus.
They also suggest that the test is not necessarily limited to bacteria. With some further research, it could also be used to detect other microbes, such as viruses. The team is now looking for ways to commercialize this innovative bacteria identification technique. Hopefully, it will take us one step ahead in our fight against pathogens.
The study is published in the journal Nano Letters.



