Study uncovers how bacteria use ancient mechanisms to self-repair
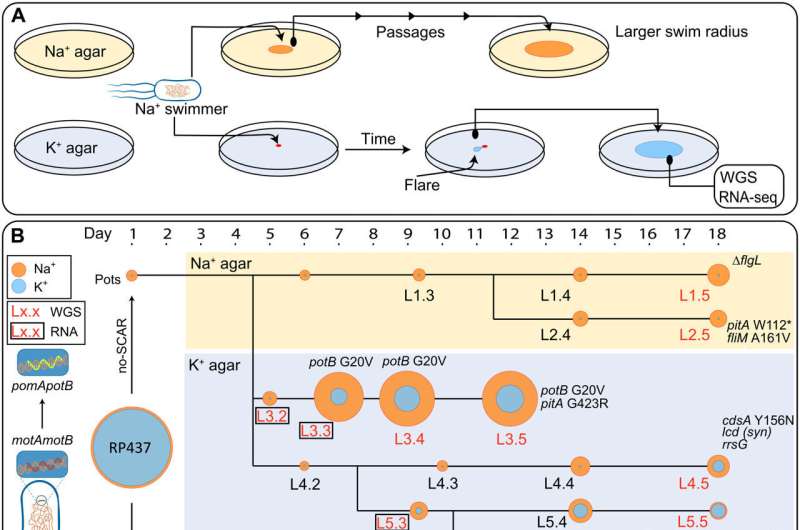
 A) Sodium-swimming strain was repeatedly passaged on either Na+LB (~100 mM [Na+]) or K+LB (~15 mM [Na+]) plates. Flares, indicating a potentially upmotile variant, were passaged and sent for sequencing. (B) Edited E. coli strains Pots and Potsλ, obtained from E. coli RP437 via no-SCAR and λ-Red recombineering, respectively, were passaged on soft agar (colored background, yellow: Na+; blue: K+) over an 18-day period, and eight lineages (L1 to L8) were selected for further investigation, each composed of five members (i.e., L1.3 indicates the first lineage and the third passage). Swimming ability in the presence of high or low sodium is displayed by a yellow or blue ring, respectively, corresponding to swim size on swim plate (C). Lack of motility on K+ soft agar is represented by a blue dot indicating colony growth only. Colonies that were nonmotile in K+ plates were confirmed with further incubation (fig. S3C). Red and boxed lineage member labels indicate WGS and RNA-seq data availability (including Pots). SNPs identified relative to the Pots reference genome are annotated next to each respective lineage member and in table S1. Highlighted genes other than pomA and potB: pitA (metal phosphate:H+ symporter), flgL (flagellar hook-filament junction protein 2), fliM (flagellar motor switch protein), cdsA (cardiolipin-diglyceride synthase), icd (isocitrate dehydrogenase), and rrsG (16S ribosomal RNA). Scale bar, 10 mm. (C) Recapitulation of the directed evolution experiment. Na+ (left) and K+ (right) soft agar plates inoculated with a 1-μl aliquot of glycerol stock of each strain indicated in (A) (except RP437) and arranged in the same order as (A). (D) Phylogeny of motB across 82 species with ancestral reconstruction at the G20 site. G20 is conserved in the Vibrio spp. clade. Full phylogeny is shown in fig. S14. Credit: Science Advances (2022). DOI: 10.1126/sciadv.abq2492″ width=”800″ height=”530″>
A) Sodium-swimming strain was repeatedly passaged on either Na+LB (~100 mM [Na+]) or K+LB (~15 mM [Na+]) plates. Flares, indicating a potentially upmotile variant, were passaged and sent for sequencing. (B) Edited E. coli strains Pots and Potsλ, obtained from E. coli RP437 via no-SCAR and λ-Red recombineering, respectively, were passaged on soft agar (colored background, yellow: Na+; blue: K+) over an 18-day period, and eight lineages (L1 to L8) were selected for further investigation, each composed of five members (i.e., L1.3 indicates the first lineage and the third passage). Swimming ability in the presence of high or low sodium is displayed by a yellow or blue ring, respectively, corresponding to swim size on swim plate (C). Lack of motility on K+ soft agar is represented by a blue dot indicating colony growth only. Colonies that were nonmotile in K+ plates were confirmed with further incubation (fig. S3C). Red and boxed lineage member labels indicate WGS and RNA-seq data availability (including Pots). SNPs identified relative to the Pots reference genome are annotated next to each respective lineage member and in table S1. Highlighted genes other than pomA and potB: pitA (metal phosphate:H+ symporter), flgL (flagellar hook-filament junction protein 2), fliM (flagellar motor switch protein), cdsA (cardiolipin-diglyceride synthase), icd (isocitrate dehydrogenase), and rrsG (16S ribosomal RNA). Scale bar, 10 mm. (C) Recapitulation of the directed evolution experiment. Na+ (left) and K+ (right) soft agar plates inoculated with a 1-μl aliquot of glycerol stock of each strain indicated in (A) (except RP437) and arranged in the same order as (A). (D) Phylogeny of motB across 82 species with ancestral reconstruction at the G20 site. G20 is conserved in the Vibrio spp. clade. Full phylogeny is shown in fig. S14. Credit: Science Advances (2022). DOI: 10.1126/sciadv.abq2492″ width=”800″ height=”530″>A new study led by UNSW Sydney scientists unveils how nature’s oldest wheel, found within bacteria, can fix itself when times get tough.
The findings, published today in Science Advances, show how the flagellar—the ancient motor that powers the swimming ability of bacteria—can also help these tiny organisms adjust to conditions where their motility is impaired.
Bacteria are one of Earths’ oldest living organisms. They are tiny single-celled organisms found across every habitat, including the human body—where there are more bacterial cells than human cells.
Being able to swim is crucial to how bacteria survive and spread. But little is known about how the motors that drive their movement help the organisms adapt to hostile environments.
The researchers from the School of Biotechnology and Biomolecular Sciences are the first in the world to use CRISPR gene-editing technology to alter a flagellar motor. They used synthetic biology techniques to engineer a sodium motor onto the genome to create a sodium-driven swimming bacteria. They then tested and tracked the bacteria’s ability to adapt when the environment was starved of sodium.
Sodium is an ion, which means that it carries a charge. It is this charge that powers the flagellar motor via stators, or ion channels.
The team found that the stators were able to rapidly self-repair the flagellar motor and restore movement. These findings could lead to new advances across the biological and medical science fields.
“We showed that environmental changes can cause ion channels to react quickly,” said lead author of the paper Dr. Pietro Ridone.
“So, the CRISPR edits also revert quickly, and the flagellar motor evolves and then regulates itself,” Dr. Ridone said.
“The fact that we saw mutations directly on the stators right away is surprising, and also inspires a lot of our future research plans in this area.”
The power of molecular machinery
The human body contains around 10,000 different types of molecular machines, which power a range of biological functions from energy conversion to movement.
The technology of a bacterial motor far surpasses what humans can synthetically engineer at nanoscale. At a millionth the size of a grain of sand, it can assemble itself and rotates at up to five times the speed of a Formula 1 engine.
“The motor that powers bacterial swimming is a marvel of nanotechnology,” said Associate Professor Matthew Baker, a co-author of the paper. “It is the absolute poster child for ancient and very sophisticated molecular machinery.”
A/Prof. Baker said the study’s findings can help us better understand the origin of molecular motors in mechanistic detail—how they came together and how do they adapt.
“These ancient parts are a powerful system to study evolution in general, as well as the origins and evolution of motility.”
A/Prof. Baker says the findings will inform how synthetic biology can help create new molecular motors. The findings may also have applications in understanding antimicrobial resistance and the virulence of disease.
“By shedding more light on life’s ancient history, we are gaining knowledge to create tools that can help better our futures,” A/Prof. Baker said. “It can also lead us to insights on how bacteria might adapt under future climate change scenarios.”
More information:
Pietro Ridone et al, The rapid evolution of flagellar ion selectivity in experimental populations of E. coli, Science Advances (2022). DOI: 10.1126/sciadv.abq2492
Provided by
University of New South Wales
Citation:
Study uncovers how bacteria use ancient mechanisms to self-repair (2022, November 23)
retrieved 23 November 2022
from https://phys.org/news/2022-11-uncovers-bacteria-ancient-mechanisms-self-repair.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.



