Graphene Oxide Decreases Pro-Inflammatory Proteins Production in Skeletal Muscle Cells Exposed to SARS-CoV-2 Spike Protein

Introduction
The research´s motivation was to explain the role of muscle tissue as a very large pool of secretory cells in the course and effects of COVID-19, particularly in the induction of cytokine storm, and the search for an effective scavenger of these pro-inflammatory proteins.
The global problem caused by the coronavirus disease (COVID-19) pandemic is the result of not only the severe, unpredictable and difficult-to-treat course of the disease, but also its incalculable consequences.1,2 A key symptom of COVID-19 is cytokine storm (CS).3 CS is manifested by excessive secretion of pro-inflammatory cytokines that initiate many pathways involved in the propagation of inflammation leading to fever, impaired capillary permeability, acute respiratory failure, multi-organ failure, and in severe cases to death.4 According to these authors, the COVID-19-related cytokine storm and its comprehensive diagnosis are the basis for the diagnosis and treatment of this disease. The key CS chemokines and cytokines are believed to include the C-X-C chemokine motif 10 (CXCL10), interferon gamma (IFN-γ), interleukin 1 beta (IL-1β), interleukins: IL-2, IL-6, IL-7, IL-8, IL-10, IL-12, IL-17, IL-18, tumor necrosis factor alpha (TNF-α), granulocyte colony-stimulating factor (G-CSF); granulocyte – macrophage colony-stimulating factor (GM-CSF) and monocyte chemoattractant protein-1 (MCP-1).4,5 Currently, COVID-19 is referred to as a multi-organ dysfunction promoting systemic inflammation.6
The most dangerous consequence of this disease is the fact that a cytokine storm can induce pro-inflammatory mechanisms in various organs and tissues over the next months and possibly years. 30–50% of patients show symptoms of the disease for many months.7 Moreover, 80% of COVID-19 infections are mild, unhospitalized and unmonitored.8 Thus, the effects of infection, including CS, may develop in a subclinical and uncontrolled manner, and serious effects may become apparent after months.7
Skeletal muscle dysfunction is also observed during and after COVID-19, and may be caused by CS induced in other tissues.9 Research is being carried out on the potential possibility of infection of muscle tissue or skeletal muscle cells by the SARS-CoV-2 virus. The expression of the TMPRSS2 and ACE2 genes in human muscle tissue was analyzed and it was confirmed that endothelial cells, smooth muscle cells, pericytes, muscle stem cells (satellite cells), macrophages, immune cells (B, T or NK cells), and muscle fibers express TMPRSS2.7,10 However, according to some authors, the ACE2 receptor is only expressed by smooth muscle cells and pericytes.8 According to other authors, skeletal muscles has high expression of the ACE2 receptor,11,12 but lower than endothelial cells.10 Yamamoto et al13 argue that there is no evidence of direct SARS-CoV-2 invasion into skeletal muscles, moreover, no MERS-CoV-2 virus particles were detected in ultrastructural studies of skeletal muscles of patients.14
Muscle tissue constitutes a significant mass of human organism, in a 50-year-old male it can constitute 36.8% of body weight,15 and on average it accounts for about 40% of total body weight, which means that it has a huge pool of various proteins, including also functional. This tissue is thought to contain 50–70% of all body proteins.16 Although muscle cells are not primarily involved in cytokine induction, however, even their minimal production, given the size of the muscle mass, can provide a powerful supply of pro-inflammatory proteins. So, paradoxically, young, athletic men may be much more sensitive to CS. On the other hand, the pain and weakness of the muscles observed during and after the disease,17 and the consequent lack of physical activity, as well as the limitation of physical activity related to quarantine also constitute the causal mechanism of CS. Therefore, the search for a method to reduce the intensity of CS is necessary.
Graphene oxide (GO) is an allotropic form of carbon made of flakes 1.1 ± 0.2 nm thick, which have the structure of graphene, but contain numerous oxygen groups such as epoxide (= O), carbonyl (C = O), hydroxyl (-OH) and on the edges carboxyl (-COOH) and carbonyl (-CO) groups.18 These numerous oxygen groups make GO hydrophilic, and after penetration into the body, it is easily passivated by various chemical compounds, especially proteins, which is called the protein corona effect.19 These features of GO also make it relatively biocompatible,20 as it is surrounded by proteins that reduce its cell-toxic effect.21 Research carried out by our team has shown that GO used as a nano-scaffold for muscle cells is not only non-toxic but is also preferentially chosen by cells that adhere and willingly inhabit the GO surface.22 Moreover, cells develop so well on the GO scaffold that they undergo spontaneous physiological contraction. Also, graphene administered to the breeding medium is non-toxic at a level below 10 μg / L.23 Studies of relatively high biocompatibility of GO have also been confirmed by us in experiments on rats,24,25 3D skin model26 and in studies on other biological models.27 It seems that GO, as a super-thin layer of carbon applied in a small amount, can, without inducing toxicity, deposit on its surface CS-related proteins present in the body. The use of such anti-inflammatory therapy could significantly reduce the dangerous, further consequences of COVID-19 disease.
The aim of the conducted experiments was to explain several important mechanisms of the COVID-19 disease and to initially attempt to use GO as an adjunct treatment.
First of all, we wanted to confirm the controversial presence of the ACE2 receptor on the surface of skeletal muscle cells, as well as determine the effects that may result from the presence of a viral fragment – the spike protein, without infecting the cell with the virus. Moreover, assuming that the S protein induces CS in the immunogenic muscle cells, it seems important which cytokines and other functional proteins would be activated. Most of all, however, for the first time, we have attempted to minimize CS by using GO. This research is the world’s first experiment on the potential possibility of GO scavenging CS proteins.
Materials and Methods
GO Characterization
A GO water solution (4mg/mL) was obtained from Advanced Graphene Products (Zielona Gora, Poland). In the experiment, dilutions were prepared in deionized water and sonicated for 15 minutes before use.
The MIR spectrum of the GO water solution was registered with KRS round plates in transmission mode. Five separate spectra of the working sample were registered, and an averaged spectrum was calculated. The Perkin Elmer System 2000 spectrometer was used for spectra registration, and analysis was performed in the Pegrams software.
The morphology of the GO flakes was examined using a TEM – JEM-1220 (JEOL, Japan) at 80 kV and a TEM CCD Morada 11 megapixels camera (Olympus Soft Imaging Solutions, Germany). The zeta potential of the GO solution was measured using a ZetaSizer Nano ZS model ZEN3500 (Malvern Instruments, UK).
Cell Culture
Human skeletal myoblast cells were purchased from Gibco (Thermo Fisher Scientific, USA). The cells were maintained in Dulbecco’s Modified Eagle’s medium (DMEM) (Life Technologies, USA) supplemented with 2% horse serum (Life Technologies, USA) and 1% penicillin/streptomycin (Life Technologies, USA) according to the manufacturer’s instructions. The cell cultures were maintained at 37°C in a humidified atmosphere of 5% CO2/95% air in a Memmert ICO150med incubator (Memmert, Germany). The Sars-COV-2 spike proteins (PX-COV-P049) were purchased from ProteoGenix (France).
The cells were divided into experimental groups: 1) control group (not treated), 2) with addition of S proteins, 3) with addition of a GO solution, 4) GO-S-with addition of GO followed by the addition of S proteins. The GO was mixed with the culture media yielding final concentrations of 100 ppm GO. In the group with GO and S proteins, a medium containing GO was applied first, and S proteins were added at a final concentration 5 µg/mL after one hour. During the experiment in the laboratory was maintained a high standard of biosafety practice, although protein S is not an infectious or replicating factor, with low/moderate individual risk and no community risk. The experiment was conducted in BSL-2 laboratory with BSL-3 biosafety protocols. No formal approval from the Institute review board was required for this study.
The cells were examined, and their morphologies were recorded using a light optical inverted microscope (TL-LED, Leica Microsystems, Germany). For further visualization, the cells were fixed in ice cold methanol for ten minutes at 4°C and stained with eosin/hematoxylin (E+H).
Confocal Microscopy
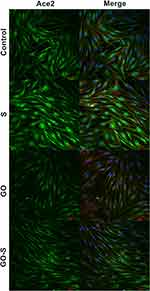
The morphology of the cells and the presence of ACE2 receptors in muscle cells was evaluated using a confocal microscope (Olympus FV1000, Japan). For imaging, the cells cultured on slides were fixed using 4% paraformaldehyde in phosphate buffered saline (PBS) (Life Technologies, USA) for ten minutes at room temperature and washed with ice-cold PBS. The cells were then incubated in Tween (0.3%) and washed with PBS (Sigma-Aldrich, USA). Subsequently, the cells were incubated with a blocking solution – normal goat serum (5%; Chemicon International, USA) – for one hour at room temperature. The cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific, USA), actin filaments were stained with phalloidin-Atto 633 (Sigma-Aldrich, USA), and the ACE2 receptors were stained with a polyclonal rabbit antibody anti-ACE2 (SAB3500978, Sigma-Aldrich, USA) overnight at 4°C (dilution 1.5:100). After three series of washing, a goat anti-rabbit antibody (dilution 1:100) was used as a secondary antibody with Alexa FluorTM 488 (Thermo Fisher Scientific, USA), and the cells were incubated for one hour at room temperature. After staining, the cells were washed with PBS, and the slides were sealed and stored in the dark.
The number of nuclei was analyzed using ImageJ-Cell Counter; the nuclei stained with DAPI were counted manually in a randomly selected field of view (n = 5/group).
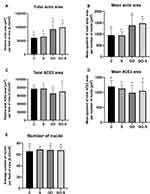
The areas of actins and ACE2 receptors were analyzed using ImageJ by calculating the actin or ACE2 positive (non-black) pixels on binarized one-channel images obtained by confocal microscopy (n = 5/group). The mean area of actins and ACE2 were obtained by dividing the total area by the number of nuclei in the field of view (n = 5/group), which represented the relative area of actins and ACE2 receptors in a single cell.
The data were analyzed using monofactorial analysis of variance, ANOVA, with Tukey’s post-hoc test for determining the differences between the groups. Differences with a p-value ≤0.05 were defined as statistically significant. Differences between groups were marked with different letters above the bars (a, b).
Analyses were performed in StatGraphics Centurion version XVI (StatPoint Technologies, USA).
Human Inflammation Antibody Array
The cells were cultured according to the scheme described in section “Cell culture” and treated for 24 hours after confluency reached about 80%. After treatment, the cells were manually detached, collected, and centrifuged at 1200 rpm for ten minutes. The protein extract was prepared with a PierceTM RIPA buffer (Thermo Scientific, USA) with protease inhibitors (Sigma-Aldrich, USA) and sonicated for three minutes with a pulse of 30 seconds on/off and an amplitude of 20%. The samples were centrifuged at 12,000 rpm for 25 minutes at 4°C. The supernatant was collected, and the total protein concentration was determined using a bicinchoninic acid kit (Sigma-Aldrich, USA). All samples from each experimental group were pooled before further procedure. Human inflammation factors were analyzed using an antibody array (ab134003; Abcam, UK). The assay was performed in accordance with the manufacturer’s instructions. The membranes were visualized using Azure Biosystem C400 (Azure, USA), and analyzed using the ImageJ software. Obtained results were normalized to the control dots.
Mass Spectrometry
Cells were cultured under standard conditions, and after confluency reached around 80%, treated for 24 hours according to the scheme described in the “Cell culture” section. After treatment, the cells were manually scrapped off, collected, washed with PBS, and centrifuged at 1200 rpm for ten minutes. The protein extract was prepared with a PierceTM RIPA buffer (Thermo Scientific, USA) and sonicated for three minutes with a pulse of 30 seconds on/off and an amplitude of 20%. Protease inhibitors (Sigma-Aldrich, USA) were added to the PierceTM RIPA buffer. The samples were centrifuged at 12,000 rpm for 25 minutes at 4°C. The supernatant was collected and subsequently precipitated using chloroform/methanol protocol. Each group was analyzed in three independent replicates. The protein pellets of each sample were solubilized in 100 µL of 100 mM TEAB, 8 M urea, 5 µL of TCEP mM, sonicated, and alkylated with 5 µL of MMTS for 30 minutes. The protein mixture was digested with a mixture of proteases LysC/Trypsin (Promega V5073, USA) in 8 M urea and 100 mM TEAB, in a 1:50 ratio. After four hours of digestion with shaking at 37°C, the samples were diluted to achieve a urea concentration of 1 M and digested overnight. The peptide mixture was purified with HL 10 mg cartridges and evaporated to dryness in a rotary evaporator. The pellets were dissolved with 2% acetonitrile in the presence of 0.1% TFA. Liquid chromatography-mass spectrometry (LC-MS) analyses were performed in the Laboratory of Mass Spectrometry (IBB PAS, Warsaw) using a NanoAcquity UPLC System (Waters) coupled to a QExactive Orbitrap Mass Spectrometer (Thermo Fisher Scientific, USA). The mass spectrometer was operated in the data-dependent MS2 mode, and data were acquired in the m/z range of 200–2000. Peptides were separated by a 180-minute linear gradient of 95% solution A (0.1% formic acid in water) to 45% solution B (acetonitrile and 0.1% formic acid). Each sample measurement was preceded by three washing runs to avoid cross-contamination.
Data were analyzed with the PEAKS Q software and searched against the UniProt human reference proteome. The results were also analyzed in the Panther Classification System version 16.0.
Heat maps were prepared using the GraphPad Prism software version 8.1.2 (GraphPad Software Inc., USA).
The data were analyzed by monofactorial analysis of variance, ANOVA, and t-test, with the level of significance set at p ≤0.05. Analyses were performed in the StatGraphics Centurion version XVI (StatPoint Technologies, USA).
Results
Characterization of GO
Transmission electron microscopy (TEM) was used to confirm the morphology of the GO flakes. After drying, thin sheets of GO formed a wrinkled surface of 1–3 layers of flakes with irregular edges (Figure 1A). The size of the flakes was 2 to 3 μm. The zeta potential of the GO water solution (1mg/mL) was −28.3 ± 2.1 mV.
 |
Figure 1 Transmission electron microscopy images of GO flakes (A); middle infrared (MIR) spectrum of graphene oxide (GO) registered in transmission mode in the spectral range of 4000–400 cm−1 (B). |
A wide characteristic band generated by the stretching of the O-H group at 3450 cm−1 was identified in middle infrared (MIR) analysis. The bands generated by the stretching of the C-H bond were located at 2816 cm−1. Bands assigned to triple bonds between carbon atoms (sp hybridization) were registered at 2363 cm−1. A very intense characteristic band at 1615 cm−1 was assigned to double bonds between carbon atoms (sp2 hybridization of carbon). Two bands located at 1371 and 1350 cm−1 were assigned to C-O or C-C stretching and/or C-H deformations, respectively. C-C stretching indicates the sp3 hybridization of carbon. Hence carbon was present in the sample in all known types of hybridization. The presence of hydrogen and oxygen was also confirmed (Figure 1B).
Cell Morphology – Light and Confocal Microscopy
The morphology of the human skeletal myoblast (HSkM) cells exposed to GO, S proteins, and S proteins together with GO were added to the culture medium in concentrations of 100 ppm GO and 5μg/mL S protein (Figure 2A and B) was analyzed using light microscopy. There were no significant differences between groups after 24 hours of treatment. An elongated shape characteristic of skeletal muscle cells was observed in all groups. The muscle fibers with multiple nuclei also were present (Figure 2B). There were no signs of apoptosis or necrosis in any of the groups. Clearly visible GO flakes were suspended in the culture medium and adhered to the cells in the GO group. Interestingly, the GO flakes adhering closely to the cells did not affect the deformation of the cells in these adhesion areas. In summary, the introduction of the S proteins and the use of GO in groups of cells incubated with S proteins, as well as the introduction of GO alone into the medium did not adversely affect the morphology of the muscle cells.
Sections with stained nuclei (DAPI), actins (phalloidin), and ACE2 receptors were observed with a confocal microscope (Figure 3), and the surface of the actins and ACE receptors were measured. No adverse changes were found in the morphological image of the actin. However, their surfaces were slightly larger in the groups where GO was applied, and in the case of total actin area in the GO-S group, it was significantly larger. However, no morphological changes indicative of cytotoxicity and no disruptions of cytoskeleton integrity or actin polymerization were observed (Figure 4A and B). The muscle cell samples were also stained for the ACE2 receptors. A strongly expressed presence of ACE2 receptors was found on muscle cells from the control group and the S, GO, and S-GO groups (Figure 3). The total ACE2 area and mean ACE2 area measurements did not differ between the groups (Figure 4C and D). When analyzing the morphology and the number of cell nuclei, no differences were observed between the groups and no pathological changes in the images of cell nuclei were found (Figure 4E).
Protein Analysis
The experiment analyzed the expression of 40 proteins involved in inflammation (Figure 5). Cells incubated with the S proteins showed an increase in ICAM-1 (A-B/9), MCP-1 (E-F/6), and IL-8 (C-D/8) proteins. The application of GO to the cells incubated with S proteins decreased the expression of these proteins to the level of the control group. Moreover, GO, when used alone, did not increase the expression of pro-inflammatory proteins.
 |
Figure 5 Array analysis of inflammation factors in HSkM cells in the control group (C) and after 24 hours of treatment with S protein (S), graphene oxide (GO), and a combination of S proteins and GO (GO-S) (A); results normalized to the control dots; full array map available in the Supplementary Material (Table S1). Abbreviations: ICAM1, intercellular adhesion molecule 1; MCP-1, monocyte chemoattractant protein 1; IL-8, interleukin 8. |
Proteomic analysis was also carried out using mass spectrometry (full data available in the Supplement Material, Table S2). We analyzed the changes in the profile of a broad spectrum of proteins induced by the presence of S proteins, GO, and both factors using the Peaks Q and Panther softwares. A total of 667 proteins with significantly altered expression levels were found in the experimental groups compared to the control group. The analysis of the heatmap images (Figure 6) shows that the S proteins increased the expression of proteins relative to the control group, while the use of GO in the group of cells incubated with S proteins decreased their level; however, the greatest decrease of numerous proteins level was observed in group treated with GO.
A volcano plot analysis confirmed these trends, showing a significant up-regulation of 13 selected proteins and a downregulation of 13 induced by the presence of S proteins in the culture media. The addition of GO to the medium of cells incubated with S proteins significantly increased the levels of five proteins and decreased the levels of 638 proteins (Figure 7). For comparison, the addition of GO alone increased the expression of five proteins and decreased the expression of 431 proteins (Table 1). The Venn diagram (Figure 8) showed number of proteins with decreased expression level, which were common in S, GO and GO-S groups, in comparison to the control group. There were no similarities in proteins with increased expression level between S group and GO, GO-S. Only one record was common between GO and GO-S group.
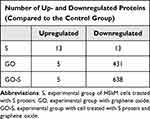 |
Table 1 Number of Proteins with Significantly Altered Expression Level Identified Through the Volcano Plot Analysis |
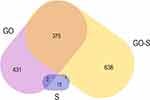 |
Figure 8 Venn diagram with numbers of proteins with significantly decreased expression level among the S, GO and GO-S groups, in comparison to the control group. |
The top ten proteins that were most significantly differentiated by the S proteins were selected (Table 2). The expression of these proteins increased to the greatest extent (at least two-fold) compared to the control group. Then, the level of these proteins was monitored following the application of GO and S proteins (GO-S group) and after using GO alone. We found that the S proteins increased the expression of the histone deacetylase 2 (HDAC2) protein the most, and interestingly, the addition of GO further increased the expression of this protein. The application of GO alone also increased its expression, but to a lesser extent. The expression of the stomatin-like protein 2 mitochondrial (STOML2) increased significantly under the influence of the S proteins, but the presence of GO was the reason for its reduction to the control group level. The S proteins also significantly increased the expression of the following proteins by more than two-fold: mitochondrial superoxide dismutase (SOD2), peroxisomal multifunctional enzyme type 2 (HSD17B4), cyclooxygenase-1 (COX-1/PTGS1), CD59 glycoprotein (CD59), calponin-3 (CNN3), caveolae-associated protein 4 (CAVIN4), titin (TTN), and 40S ribosomal protein S3 (RPS3). Interestingly, in all cases, application of GO decreased the level of these proteins. The incubation of cells with S proteins also resulted in a significant, more than two-fold reduction in the expression of two proteins – keratin type I cytoskeletal 9 (KRT9) and hornerin (HRNR).
The application of GO significantly increased the expression of these proteins, and indeed, the level of these proteins was the highest in the groups with GO alone.
Regarding the influence of GO, the levels of several proteins increased relative to the control group, especially keratin type II cytoskeletal 2, hornerin, and keratin type II cytoskeletal. The level of the remaining proteins was lower than in the control group.
Around one third of proteins were responsible for cellular processes and about 1/5 for metabolic changes (Figure 9A). With regard to molecular functions, approximately 45% of the different proteins were related to binding and as much as 30% were associated with catalytic activity (Figure 9B).
Discussion
A Human Skeletal Myoblast (HSkM) was derived from a healthy human being and constituted the physiologically relevant cellular model of myoblasts. Cells taken directly from muscle tissue that have undergone few population doublings and have not been immortalized represent a more reliable experimental model of skeletal muscle response to experimental factors compared to traditionally used immortalized cells.28
In studies carried out on HSkM cells, we found a well-expressed presence of the ACE2 receptor protein localized around the cells, which is confirmed by the studies of other authors.11,29 The expression of the ACE2 receptor in muscle cells indicates the potential effect of SARS-CoV-2 on skeletal muscle17,30 and may explain the cause of muscle weakness, pain and dysfunction during COVID-19 disease.31 There is a significant loss of muscle mass in COVID-19 patients.32 Among the studies documenting skeletal muscle damage related to COVID-19, the authors point to sarcopenia,33 cachexia, myalgia, myositis, rhabdomyolysis, atrophy, peripheral neuropathy and Guillain-Barré syndrome.32–35 In our research, we did not observe any pathological changes in the morphological image, especially the cell microstructure, the number of nuclei and the actin image. However, in our experiments we did not use the SARS-CoV-2 virus, but only its S spike protein. Protein S is a key determinant of the invasiveness of the virus by binding to ACE2, hence the inhibition of the SARS-CoV-2 S – ACE2 interaction is the basis for research on the treatment and prevention of COVID-19.36,37 In our research, we observed a change in the spectrum of proteins produced by muscle cells under short contact with the spike protein. This should be related to the situation where there is no infection of muscle cells but only contact with the residues of the virus. Moreover, in vitro studies on one cell line never reflect the real state, where the condition of muscles is also influenced by proteins and secretions synthesized in other tissues. In vitro studies on primary cells are carried out in a short period of time due to the mortality of HSkM cells. However, these studies are crucial in understanding the first signs of a cytokine storm that may occur under the influence of the S protein in muscle tissue. According to Seixas et al38 the mechanisms responsible for the pathological changes in skeletal muscles during COVID-19 are still unknown, but undoubtedly the key cause, according to these authors, seems to be a cytokine storm.
Thus, another study of the implications that might be associated with the presence of the viral spike protein was to determine the expression of proteins involved in the induction of inflammation. After incubation of HSkM cells with the S protein, an increase in the levels of ICAM-1, MCP-1 and IL-8, which are among the key pro-inflammatory cytokines and chemokines, was observed.39 Increased levels of these proteins have also been observed in COVID-19 patients.4 Interleukin 8 is a chemokine considered to be the most potent chemotactic agent in humans, and moreover has a clear target specificity for neutrophils, which it attracts and activates in inflammatory areas.40 Muscle tissue shows the presence of many cytokines and chemokines, including IL-8,41 which is a manifestation of its immunological and endocrine function.42 However, some authors argue that a modest release of IL-8 by muscle cells causes not so much an increase in plasma IL-8 concentration as local activity, especially promoting angiogenesis.42,43 IL-8 may bind the CXCR1 receptor responsible for its pro-inflammatory action or the CXCR2 receptor present on microvascular endothelial cells responsible for angiogenesis.44 This local proangiogenic neovascularization promoting effect of IL-842 may, paradoxically in the case of COVID-19, increase the ACE2 receptor pool present on the endothelial cells of newborn vessels and thus increase the cell availability for SARS-CoV-2 virus.
Another protein whose elevated level was observed due to the presence of the S protein was the chemokine MCP-1. MCP-1 is responsible for the production of adhesion molecules as well as the proliferation and migration of VSMC vascular smooth muscle cells45,46 and may also promote the formation of atherosclerotic lesions, it is one of the most important cytokines responsible for venous thrombosis.47 In COVID-19 patients, a postmortem examination revealed thrombus in the small vessels of the lungs. The presence of a fibrin clot in small arteries indicates an increased risk of vascular and venous thromboembolic complications.48,49 Moreover, serum MCP-1 levels are a biomarker indicating a very severe course and a high risk of death in COVID-19 patients.5 Studies on mice have found that increased production of MCP-1 in skeletal muscle promotes inflammation in skeletal muscle.50 Moreover, MCP-1 is also called myokine – the exercise factor, because intense muscle effort increases its level, and it promotes the infiltration of macrophages after severe muscle damage. It appears that muscle, as a tissue that exhibits secretory functions and accounts for approximately 40% of body weight, may play a very important role in inducing and promoting inflammation, especially in COVID-19.
In the conducted studies, an increased level of ICAM-1 was also observed under the influence of the spike S protein. Increased blood ICAM-1 levels have been reported in patients with COVID-19, especially in severe cases, and ICAM-1 may also be a predictor of complications related to COVID-19.51 ICAM-1 is a cell surface adhesion receptor and is responsible for the accumulation of white blood cells at sites of inflammation. Moreover, it stimulates the synthesis of cytokines that induce rhinovirus replication.52 Increased ICAM-1 expression in muscle cells and muscle satellite cells occurs after muscle overload, contributing to muscle hypertrophy.53 Exercise may also increase the expression of IL-8, increased levels of which were observed under the influence of the S protein.54 Thus, the spike protein is likely to induce mechanisms similar to severe muscle overload. Muscle overload reduces the antioxidant capacity, which can lead to oxidative stress.55 Activation of oxidative stress can be a step in the development of a cytokine storm. Analysis of the level of cytokines and the found increased synthesis of IL-8, MCP-1 and ICAM-1 – characteristic of COVID-19 infection,56 induced in human skeletal muscle cells by the presence of the spike protein, clearly indicated the activation of the cytokine storm.
The application of GO in the group of cells incubated in the medium with the S protein decreased the level of cytokines IL-8, MCP-1 and ICAM-1 to the level of the control group. Graphene oxide alone, added to the culture medium, did not affect the induction of any of the inflammatory proteins. These results confirm ours and other authors’ observations, which prove the significant biocompatibility of graphene oxide administered in a small amount.23 In in vitro studies on smooth muscle, found no negative effect of GO, but it was observed an increase of cell proliferation under the influence of GO.57 Moreover, in vitro studies on macrophages showed that GO at 1 µm / mL did not increase the level of pro-inflammatory cytokines IL-6 and TNF-α.58 There are also studies showing negative, including pro-inflammatory effects of GO,59 although the toxicity depends on the dose used and we used a safe level in our studies, also verified in preliminary studies.
In order to clarify the molecular causes of changes induces by the incubation of cells with the S protein and the effect of GO to reduce this adverse effect, we conducted a proteomic analysis of the lysate of cells from the control group and the S, GO-S and GO groups. We found that the S protein had the greatest effect on increasing the level of histone deacetylase (HDAC2), what’s more, GO increased the level of this protein even more. Takahashi et al60 indicate that HDAC2 increases ACE2 expression, being an important risk factor in the course of COVID-19. This is very clearly confirmed by our results, which moreover documented that the S protein, regardless of the presence of a virus, causes this mechanism in skeletal muscle cells. It can be assumed that the spike protein promotes infection by increasing the ACE2 pool and thus increasing the availability of cells to the virus. ACE2, in addition to its function of cleaving angiotensin II, also affects the functioning of mitochondria,61 what is more, it regulates the activity of the mitochondrial oxidase NADPH 4, responsible for the production of Reactive Oxygen Forms (ROS).62 The overproduction of ROS may, in turn, be a factor that promotes a cytokine storm.
Another protein whose level has significantly increased is Stomatin-like protein 2 mitochondrial (STOML2). This protein regulates the biogenesis and activity of mitochondria by regulating mitochondrial translation, although its key role is to increase the activity of this translation in a state of increased cell activity.63 It can be presumed that the increase in HDAC2 and STOML2 levels is associated with the activation of the ROS production process by the S protein. This may be confirmed by a significant increase in the level of Superoxide dismutase (SOD2), Cyclooxygenase-1 (COX-1) and Peroxisomal multifunctional enzyme type 2 (HSD17B4) and the 40S S3 ribosomal protein (RPS3). SOD2 is an enzyme that catalyzes superoxide dismutation in the first line of ROS formation in mitochondria.64 SOD2 levels may be elevated due to overproduction of ROS.65 What’s more, increasing the level of the 40S S3 ribosomal protein, which, apart from involvement in translation, also performs a repair function towards DNA, and in particular has an affinity for the DNA adduct (7,8-dihydro-8-oxoguanine (8-oxoG), resulting from the action of ROS66 would confirm the thesis about the promotion of oxidative stress in skeletal muscle cells by the spike protein.
Overexpression of SOD2 also affects the aconitase activity and may inhibit the activity of pyruvate carboxylase – the Krebs cycle enzyme, which reduces the efficiency of energy production from glucose,67 which in turn may affect the activation of beta-oxidation of fatty acids, manifested by increased PMFP2 expression. Muscle weakness is a characteristic symptom of COVID-19 infection.17 COX-1 and COX-2 are highly expressed in inflamed tissues, including COVID-19.68 COX-1 is present in muscles69 and is a physiologic variant of COX enzyme which catalyzes the conversion of arachidonic acid to Prostaglandin H2 (PGH2), leading to the synthesis of prostaglandins and tromboxan, and above all to PGG2.70 Moreover, COX-1 regulates angiogenesis,71 which would seem consistent with the increase in IL-8 levels observed in our research and support the hypothesis of promoting infection by the spike protein by activating angiogenesis and thereby increasing the ACE2 receptor pool. However, confirmation of this thesis requires further in-depth research. Nevertheless, both an increase in SOD2 and COX-1 levels may indicate activation of oxidative stress, which is a cytokine storm promoter by activating the redox-sensitive transcription factor NF-kB, which will regulate IL-1β, IL-6, TNF-α and observed in our study, IL-8.72
The S protein also increased the expression of the CD59 membrane glycoprotein, which may also be involved in the regulation of inflammatory factors.73 There was also an increase in the level of CAVIN4 – a protein significantly involved in inflammatory signaling mechanisms related to pathogenic changes in muscles, including muscular dystrophy.74 Interestingly, the S protein also increased CNN3 protein expression. This protein regulates the organization and contractility of stress fibers,75 which may suggest some influence of the spike protein on muscle function. This would confirm an increase in the level of titin, a protein strongly associated with muscle activation and strength,76 which may also confirm the observation regarding the increase in ICAM-1 expression and support the thesis that the effect of the S protein is similar to the effect of muscle overload.
Analyzing the influence of GO on the expression of the discussed proteins increased by the spike protein, it should be stated that all these proteins, except HDAC2, were down-regulated. The decreased expression of these proteins was non-specific and showed no physiological relationship. The protein corona effect characteristic of GO77 may explain this phenomenon. GO, as a super-thin material, has a large specific surface, which exposes numerous hydroxyl, epoxy and carboxyl groups, enabling the formation of weak bonds, electrostatic and hydrophobic interactions.78,79 GO flakes introduced into the biological fluid are surrounded by proteins present in the biological environment, although this process is not strictly specific, there is a certain personalization characteristic of the biological fluid donor’s disease.80,81 This regularity explains why the proteome of muscle cells treated with spike protein and GO is different compared to cells treated with GO alone. By analyzing the proteomic results, as well as the identification of selected inflammatory cytokines, it can be assumed that GO flakes bound proteins involved in promoting cytokine storm on the surface. These preliminary results represent the first attempt to use GO as a quencher for a spike protein-induced cytokine storm. The use of GO as a factor reducing the effect of CS could be of key importance for post-COVID-19 therapy.
Conclusion
It can be concluded that human muscle cells have a strongly expressed ACE2 receptor, which confirms the results of studies by other authors. This observation confirms the possibility of the SARS-CoV-2 virus entering muscle cells and stimulating them to produce pro-inflammatory proteins. In addition, the S protein of the SARS-CoV-2 virus, introduced into the muscle cell culture, activates some molecular mechanisms leading to the induction of a cytokine storm. Molecular changes under the influence of protein S may resemble the effect of muscle overload, especially the activation of oxidative stress and pro-angiogenic effects. This confirms the secretory function of skeletal muscle tissue and, for the first time, indicates the pro-inflammatory potential of this tissue in the face of COVID-19 infection. Moreover, this effect is independent of the presence of the virus in the cell, but only induced by the spike protein. Most importantly, however, we documented for the first time the possibility of reducing pro-inflammatory proteins, induced in the cell by protein S, with GO. As highly capable of adsorbing proteins on their surface and forming a protein corona, GO flakes can act as a CS protein scavenger. The conducted research may indicate the potential use of GO in supporting the treatment of muscle inflammation in COVID-19. Since muscles are a large part of the human body mass, their cytokine storm-promoting activity is likely to determine the course of the disease.
Acknowledgments
This manuscript is a part of Jaśmina Bałaban’s PhD thesis.
Funding
This research was funded by the National Center for Research and Development in Poland, project dedicated to COVID-19 Hospitals number 23/2020.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gonzalez A, Orozco-Aguilar J, Achiardi O, Simon F, Cabello-Verrugio C. SARS-CoV-2/renin-angiotensin system: deciphering the clues for a couple with potentially harmful effects on skeletal muscle. Int J Mol Sci. 2020;21(21):7904. doi:10.3390/ijms21217904
2. Rabbani G, Ahn SN. Review: roles of human serum albumin in prediction, diagnoses and treatment of COVID-19. Int J Biol Macromol. 2021;193(Pt A):948–955. doi:10.1016/j.ijbiomac.2021.10.095
3. Pum A, Ennemoser M, Adage T, Kungl AJ. Cytokines and chemokines in SARS-CoV-2 infections-therapeutic strategies targeting cytokine storm. Biomolecules. 2021;11(1):91. doi:10.3390/biom11010091
4. Yang L, Xie X, Tu Z, Fu J, Xu D, Zhou Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct Target Ther. 2021;6(1):255. doi:10.1038/s41392-021-00679-0
5. Chen Y, Wang J, Liu C, et al. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Mol Med. 2020;26(1):97. doi:10.1186/s10020-020-00230-x
6. Long B, Carius BM, Chavez S, et al. Clinical update on COVID-19 for the emergency clinician: presentation and evaluation. Am J Emerg Med. 2022;54:46–57.
7. Meacci E, Pierucci F, Garcia-Gil M. Skeletal muscle and COVID-19: the potential involvement of bioactive sphingolipids. Biomedicines. 2022;10(5):117.
8. Disser NP, De Micheli AJ, Schonk MM, et al. Musculoskeletal consequences of COVID-19. J Bone Joint Surg Am. 2020;102(14):1197–1204.
9. Suh J, Amato AA. Neuromuscular complications of coronavirus disease-19. Curr Opin Neurol. 2021;34(5):669–674.
10. Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. 2020;16(7):e9610. doi:10.15252/msb.20209610
11. Riquelme C, Acuña MJ, Torrejón J, et al. ACE2 is augmented in dystrophic skeletal muscle and plays a role in decreasing associated fibrosis. PLoS One. 2014;9(4):e93449. doi:10.1371/journal.pone.0093449
12. Perez-Valera M, Martinez-Canton M, Gallego-Selles A, et al. Angiotensin-converting enzyme 2 (SARS-CoV-2 receptor) expression in human skeletal muscle. Scand J Med Sci Sports. 2021;31(12):2249–2258. doi:10.1111/sms.14061
13. Yamamoto K, Takeshita H, Rakugi H. ACE2, angiotensin 1-7 and skeletal muscle: review in the era of COVID-19. Clin Sci. 2020;134(22):3047–3062. doi:10.1042/CS20200486
14. Alsaad KO, Hajeer AH, Al Balwi M, et al. Histopathology of Middle East respiratory syndrome coronovirus (MERS-CoV) infection – clinicopathological and ultrastructural study. Histopathology. 2018;72(3):516–524. doi:10.1111/his.13379
15. Gallagher D, Visser M, De Meersman RE, et al. Appendicular skeletal muscle mass: effects of age, gender, and ethnicity. J Appl Physiol. 1997;83(1):229–239. doi:10.1152/jappl.1997.83.1.229
16. Frontera WR, Ochala J. Skeletal muscle: a brief review of structure and function. Calcif Tissue Int. 2015;96(3):183–195. doi:10.1007/s00223-014-9915-y
17. Paliwal VK, Garg RK, Gupta A, Tejan N. Neuromuscular presentations in patients with COVID-19. Neurol Sci. 2020;41(11):3039–3056. doi:10.1007/s10072-020-04708-8
18. Schniepp HC, Li JL, McAllister MJ, et al. Functionalized single graphene sheets derived from splitting graphite oxide. J Phys Chem B. 2006;110(17):8535–8539. doi:10.1021/jp060936f
19. Maiolo D, Del Pino P, Metrangolo P, Parak WJ, Baldelli Bombelli F. Nanomedicine delivery: does protein Corona route to the target or off road? Nanomedicine. 2015;10(21):3231–3247. doi:10.2217/nnm.15.163
20. Kurantowicz N, Sawosz E, Halik G, et al. Toxicity studies of six types of carbon nanoparticles in a chicken-embryo model. Int J Nanomedicine. 2017;12:2887–2898. doi:10.2147/IJN.S131960
21. Hu W, Peng C, Lv M, et al. Protein Corona-mediated mitigation of cytotoxicity of graphene oxide. ACS Nano. 2011;5(5):3693–3700. doi:10.1021/nn200021j
22. Bałaban J, Wierzbicki M, Zielińska M, et al. Effects of graphene oxide nanofilm and chicken embryo muscle extract on muscle progenitor cell differentiation and contraction. Molecules. 2020;25(8). doi:10.3390/molecules25081991
23. Bałaban J, Zielińska M, Wierzbicki M, et al. Effect of muscle extract and graphene oxide on muscle structure of chicken embryos. Animals. 2021;11(12):1–18. doi:10.3390/ani11123467
24. Kurantowicz N, Strojny B, Sawosz E, et al. Biodistribution of a high dose of diamond, graphite, and graphene oxide nanoparticles after multiple intraperitoneal injections in rats. Nanoscale Res Lett. 2015;10(1):398. doi:10.1186/s11671-015-1107-9
25. Strojny B, Kurantowicz N, Sawosz E, et al. Long term influence of carbon nanoparticles on health and liver status in rats. PLoS One. 2015;10(12):1–19. doi:10.1371/journal.pone.0144821
26. Zielińska-Górska M, Sawosz E, Sosnowska M, et al. Molecular biocompatibility of a silver nanoparticle complex with graphene oxide to human skin in a 3D epidermis in vitro model. Pharm. 2022;14(7):116.
27. Jaworski S, Strojny-Cieślak B, Wierzbicki M, et al. Comparison of the toxicity of pristine graphene and graphene oxide, using four biological models. Mater. 2021;14(15):4250. doi:10.3390/ma14154250
28. Tan GW, Kondo T, Imamura K, et al. Simple derivation of skeletal muscle from human pluripotent stem cells using temperature-sensitive Sendai virus vector. J Cell Mol Med. 2021;25(20):9586–9596. doi:10.1111/jcmm.16899
29. Motta-Santos D, Dos Santos RAS, Oliveira M, et al. Effects of ACE2 deficiency on physical performance and physiological adaptations of cardiac and skeletal muscle to exercise. Hypertens Res. 2016;39(7):506–512. doi:10.1038/hr.2016.28
30. He Q, Sudibya HG, Yin Z, et al. Centimeter-long and large-scale micropatterns of reduced graphene oxide films: fabrication and sensing applications. ACS Nano. 2010;4:3201. doi:10.1021/nn100780v
31. Ferrandi PJ, Alway SE, Mohamed JS. The interaction between SARS-CoV-2 and ACE2 may have consequences for skeletal muscle viral susceptibility and myopathies. J Appl Physiol. 2020;129(4):864–867. doi:10.1152/japplphysiol.00321.2020
32. Ali AM, Kunugi H. Skeletal muscle damage in COVID-19: a call for action. Medicina. 2021;57(4):372. doi:10.3390/medicina57040372
33. Piotrowicz K, Gąsowski J, Michel JP, Veronese N. Post-COVID-19 acute sarcopenia: physiopathology and management. Aging Clin Exp Res. 2021;33(10):2887–2898. doi:10.1007/s40520-021-01942-8
34. Ramani SL, Samet J, Franz CK, et al. Musculoskeletal involvement of COVID-19: review of imaging. Skeletal Radiol. 2021;50(9):1763–1773. doi:10.1007/s00256-021-03734-7
35. Caress JB, Castoro RJ, Simmons Z, et al. COVID-19-associated Guillain-Barré syndrome: the early pandemic experience. Muscle Nerve. 2020;62(4):485–491. doi:10.1002/mus.27024
36. Rabbani G, Ahn SN, Kwon H, Ahmad K, Choi I. Penta-peptide ATN-161 based neutralization mechanism of SARS-CoV-2 spike protein. Biochem Biophys Reports. 2021;28:101170. doi:10.1016/j.bbrep.2021.101170
37. Angeli F, Reboldi G, Trapasso M, Zappa M, Spanevello A, Verdecchia P. COVID-19, vaccines and deficiency of ACE(2) and other angiotensinases. Closing the loop on the “Spike effect”. Eur J Intern Med. 2022;103:23–28. doi:10.1016/j.ejim.2022.06.015
38. Seixas ML, Mitre LP, Shams S, et al. Unraveling muscle impairment associated with COVID-19 and the role of 3D culture in its investigation. Front Nutr. 2022;9:825629. doi:10.3389/fnut.2022.825629
39. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi:10.1016/S0140-6736(20)30183-5
40. Bickel M. The role of interleukin-8 in inflammation and mechanisms of regulation. J Periodontol. 1993;64(5 Suppl):456–460.
41. De Rossi M, Bernasconi P, Baggi F, de Waal Malefyt R, Mantegazza R. Cytokines and chemokines are both expressed by human myoblasts: possible relevance for the immune pathogenesis of muscle inflammation. Int Immunol. 2000;12(9):1329–1335. doi:10.1093/intimm/12.9.1329
42. Akerstrom T, Steensberg A, Keller P, Keller C, Penkowa M, Pedersen BK. Exercise induces interleukin-8 expression in human skeletal muscle. J Physiol. 2005;563(2):507–516. doi:10.1113/jphysiol.2004.077610
43. Heidemann J, Ogawa H, Dwinell MB, et al. Angiogenic effects of interleukin 8 (CXCL8) in human intestinal microvascular endothelial cells are mediated by CXCR2. J Biol Chem. 2003;278(10):8508–8515. doi:10.1074/jbc.M208231200
44. Addison CL, Daniel TO, Burdick MD, et al. The CXC chemokine receptor 2, CXCR2, is the putative receptor for ELR + CXC chemokine-induced angiogenic activity. J Immunol. 2000;165(9):5269–5277. doi:10.4049/jimmunol.165.9.5269
45. van der Meer IM, de Maat MPM, Bots ML, et al. Inflammatory mediators and cell adhesion molecules as indicators of severity of atherosclerosis: the Rotterdam study. Arterioscler Thromb Vasc Biol. 2002;22(5):838–842. doi:10.1161/01.ATV.0000016249.96529.B8
46. Xiang S, Dong N, Liu J, et al. Inhibitory effects of suppressor of cytokine signaling 3 on inflammatory cytokine expression and migration and proliferation of IL-6/IFN-γ-induced vascular smooth muscle cells. J Huazhong Univ Sci Technol. 2013;33(5):615–622. doi:10.1007/s11596-013-1168-x
47. Li YS, Shyy YJ, Wright JG, Valente AJ, Cornhill JF, Kolattukudy PE. The expression of monocyte chemotactic protein (MCP-1) in human vascular endothelium in vitro and in vivo. Mol Cell Biochem. 1993;126(1):61–68. doi:10.1007/BF01772208
48. Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18(4):844–847. doi:10.1111/jth.14768
49. Perico L, Benigni A, Casiraghi F, Ng LFP, Renia L, Remuzzi G. Immunity, endothelial injury and complement-induced coagulopathy in COVID-19. Nat Rev Nephrol. 2021;17(1):46–64. doi:10.1038/s41581-020-00357-4
50. Evers-van Gogh IJA, Oteng AB, Alex S, et al. Muscle-specific inflammation induced by MCP-1 overexpression does not affect whole-body insulin sensitivity in mice. Diabetologia. 2016;59(3):624–633. doi:10.1007/s00125-015-3822-2
51. Smith-Norowitz TA, Loeffler J, Norowitz YM, Kohlhoff S. Intracellular adhesion molecule-1 (ICAM-1) levels in convalescent COVID-19 serum: a case report. Ann Clin Lab Sci. 2021;51(5):730–734.
52. Liu H, Tan J, Liu J, Feng H, Pan D. Altered mast cell activity in response to rhinovirus infection provides novel insight into asthma. J Asthma. 2020;57(5):459–467.
53. Dearth CL, Goh Q, Marino JS, et al. Skeletal muscle cells express ICAM-1 after muscle overload and ICAM-1 contributes to the ensuing hypertrophic response. PLoS One. 2013;8(3):e58486. doi:10.1371/journal.pone.0058486
54. Della Gatta PA, Garnham AP, Peake JM, Cameron-Smith D. Effect of exercise training on skeletal muscle cytokine expression in the elderly. Brain Behav Immun. 2014;39:80–86.
55. Palazzetti S, Richard MJ, Favier A, Margaritis I. Overloaded training increases exercise-induced oxidative stress and damage. Can J Appl Physiol. 2003;28(4):588–604.
56. Ebihara T, Matsumoto H, Matsubara T, et al. Resistin associated with cytokines and endothelial cell adhesion molecules is related to worse outcome in COVID-19. Front Immunol. 2022;13:830061.
57. Ren J, Braileanu G, Gorgojo P, et al. On the biocompatibility of graphene oxide towards vascular smooth muscle cells. Nanotechnology. 2020;32(5):55101.
58. Cicuéndez M, Casarrubios L, Barroca N, et al. Benefits in the macrophage response due to graphene oxide reduction by thermal treatment. Int J Mol Sci. 2021;22:13.
59. Wen KP, Chen YC, Chuang CH, Chang HY, Lee CY, Tai NH. Accumulation and toxicity of intravenously-injected functionalized graphene oxide in mice. J Appl Toxicol. 2015;35(10):1211–1218.
60. Takahashi Y, Hayakawa A, Sano R, et al. Histone deacetylase inhibitors suppress ACE2 and ABO simultaneously, suggesting a preventive potential against COVID-19. Sci Rep. 2021;11(1):3379.
61. Shi TT, Yang FY, Liu C, et al. Angiotensin-converting enzyme 2 regulates mitochondrial function in pancreatic β-cells. Biochem Biophys Res Commun. 2018;495(1):860–866.
62. Graham KA, Kulawiec M, Owens KM, et al. NADPH oxidase 4 is an oncoprotein localized to mitochondria. Cancer Biol Ther. 2010;10(3):223–231.
63. Mitsopoulos P, Lapohos O, Weraarpachai W, Antonicka H, Chang YH, Madrenas J. Stomatin-like protein 2 deficiency results in impaired mitochondrial translation. PLoS One. 2017;12:e0179967.
64. Kitada M, Xu J, Ogura Y, Monno I, Koya D. Manganese superoxide dismutase dysfunction and the pathogenesis of kidney disease. Front Physiol. 2020;11:755.
65. Warner BB, Stuart L, Gebb S, Wispé JR. Redox regulation of manganese superoxide dismutase. Am J Physiol. 1996;271(1 Pt 1):L150–8.
66. Kim SH, Lee JY, Kim J. Characterization of a wide range base-damage-endonuclease activity of mammalian rpS3. Biochem Biophys Res Commun. 2005;328(4):962–967.
67. Kim KH, Rodriguez AM, Carrico PM, Melendez JA. Potential mechanisms for the inhibition of tumor cell growth by manganese superoxide dismutase. Antioxid Redox Signal. 2001;3(3):361–373.
68. Fokunang C. Overview of non-steroidal anti-inflammatory drugs (nsaids) in resource limited countries. MOJ Toxicol. 2018;4(1):5–13.
69. Weinheimer EM, Jemiolo B, Carroll CC, et al. Resistance exercise and cyclooxygenase (COX) expression in human skeletal muscle: implications for COX-inhibiting drugs and protein synthesis. Am J Physiol Regul Integr Comp Physiol. 2007;292(6):R2241–8.
70. Williams CS, Mann M, DuBois RN. The role of cyclooxygenases in inflammation, cancer, and development. Oncogene. 1999;18(55):7908–7916.
71. Gupta RA, Tejada LV, Tong BJ, et al. Cyclooxygenase-1 is overexpressed and promotes angiogenic growth factor production in ovarian cancer. Cancer Res. 2003;63(5):906–911.
72. Attiq A, Yao LJ, Afzal S, Khan MA. The triumvirate of NF-κB, inflammation and cytokine storm in COVID-19. Int Immunopharmacol. 2021;101(Pt B):108255.
73. Huang Y, Qiao F, Abagyan R, Hazard S, Tomlinson S. Defining the CD59-C9 binding interaction. J Biol Chem. 2006;281(37):27398–27404.
74. Chidlow JHJ, Sessa WC. Caveolae, caveolins, and cavins: complex control of cellular signalling and inflammation. Cardiovasc Res. 2010;86(2):219–225.
75. Ciuba K, Hawkes W, Tojkander S, et al. Calponin-3 is critical for coordinated contractility of actin stress fibers. Sci Rep. 2018;8(1):17670.
76. Monroy JA, Powers KL, Gilmore LA, Uyeno TA, Lindstedt SL, Nishikawa KC. What is the role of titin in active muscle? Exerc Sport Sci Rev. 2012;40(2):73–78.
77. Duan G, Kang S, Tian X, et al. Protein Corona mitigates the cytotoxicity of graphene oxide by reducing its physical interaction with cell membrane. Nanoscale. 2015;7(37):15214–15224.
78. Chung C, Kim YK, Shin D, Ryoo SR, Hong BH, Min DH. Biomedical applications of graphene and graphene oxide. Acc Chem Res. 2013;46(10):2211–2224.
79. Wang M, Gustafsson OJR, Siddiqui G, et al. Human plasma proteome association and cytotoxicity of nano-graphene oxide grafted with stealth polyethylene glycol and poly(2-ethyl-2-oxazoline). Nanoscale. 2018;10(23):10863–10875.
80. Hajipour MJ, Raheb J, Akhavan O, et al. Personalized disease-specific protein Corona influences the therapeutic impact of graphene oxide. Nanoscale. 2015;7(19):8978–8994.
81. Di Santo R, Digiacomo L, Quagliarini E, et al. Personalized graphene oxide-protein corona in the human plasma of pancreatic cancer patients. Front Bioeng Biotechnol. 2020;8:491.