Incorporation of Targeting Biomolecule Improves Interpolymer Complex-Superparamagnetic Iron Oxide Nanoparticles Attachment to and Activation of T2 MR Signals in M2 Macrophages

Introduction
Inflammation is an important process in recovering homeostasis after injury or in response to a pathogen; however, it is also often indicative of or a contributor to chronic disease pathology. Inflammation is complex, comprising many cell types, signaling molecules, and cellular responses. The key contributors to inflammation are the production of reactive oxygen species (ROS) and the presence of macrophages. ROS are vital to the initiation and progression of inflammatory conditions, while macrophages are cells belonging to the innate immune system that have a high level of plasticity as they can change their phenotype in response to specific environmental stimuli.1–3 Detection of inflammation using a technique with clinically relevant spatial resolution can aid in disease diagnosis, staging, and patient-specific treatment.
When selectively activated under oxidative conditions, interpolymer complexed superparamagnetic iron oxide nanoparticles (IPC-SPIOs) affect the local T2 MR signal; thereby acting as a contrast agent that is activatable by a key component of inflammation. IPC-SPIOs are comprised of superparamagnetic iron oxide nanoparticles (SPIOs) coated in poly(ethylene glycol) (PEG), which are complexed through hydrogen bonding with poly(gallol).4–6 One of the main challenges in nanomedicine is selectively and specifically delivering nanoparticles to desired sites, which limits the translation of innovations from laboratory to clinical settings.7–12 An effective targeted nanoparticle avoids off-target effects while improving specific targeting to the disease location. In this study, mannose was incorporated into IPC-SPIOs to target receptors on M2 macrophages to allow the detection of oxidative species located in or near cells that are highly relevant to inflammatory disease diagnosis.
Mannose is a monomer of carbohydrate belonging to the aldohexose group.13 Mannose receptor is a C-type lectin that is present on M2 macrophages, antigen presenting cells, and immature dendritic cells.14 On these cells, the extracellular region’s N-terminal cysteine-rich (CR) domain binds glycoproteins bearing sulfated sugars and eight carbohydrate recognition domains,15,16 which allow high-affinity binding of mannose and fructose. The basic function of the mannose receptor is to recognize carbohydrates present on the surface of pathogens and facilitate mannose receptor-mediated endocytosis and phagocytosis.17 Other immune cells such as sinusoidal lining cells in the spleen, Kupffer cells in the liver, alveolar macrophages, and immune cells near the tumor microenvironment also express mannose receptors abundantly.
Macrophages are actively involved in inflammatory diseases, and they play a key role in initiating, maintaining, and resolving inflammation. There are several subtypes of macrophages depending on their activation state and function. The most prominent subtypes are M1 classically activated macrophages and M2 alternatively activated macrophages, which have different functions at inflammatory sites.18 M1 macrophages are pro-inflammatory and secrete factors that initiate and maintain inflammation, while M2 macrophages are anti-inflammatory and signal the transition to the resolution of inflammation. M2 macrophages are the most dominant subtype of macrophages during resolution of inflammation, rebuilding of extracellular matrix, and tumor growth. The tumor promoting capabilities of M2 macrophages involve regulating immuno-suppression, angiogenesis, neovascularization, stromal activation, and remodeling.19–21 Specifically, M2b subset also known as regulatory macrophages regulates significant portions of immune response and inflammatory reaction.22,23 M2b can promote several cancers and infectious diseases by blunting the immune and inflammatory response to tumor growth and pathogens.24–26 Recovery from injuries such as spinal cord injury and myocardial ischemia/reperfusion is also regulated by M2 macrophages.27,28 They actively regulate inflammatory diseases and facilitate the transition from pro-inflammatory conditions to resolution. M1 and M2 macrophages express some distinct phenotypic markers that can be used to identify each lineage: CD80, CD86, CD64, and CD32 for M1, while M2 macrophages express CD206 (mannose receptor 1), CD163, and CD68.18
In this study, mannose-targeted IPC-SPIOs were created to target M2 macrophages with high relevance to the detection of inflammatory diseases such as cardiovascular disease, spinal cord injury, and several cancers. We hypothesized that due to the high affinity of mannose to the mannose receptor, mannose-IPC-SPIOs would accumulate on M2 macrophages at a higher rate compared to untargeted nanoparticles. Therefore, in this study, we investigated the surface modification of IPC-SPIOs with mannose, mannose-IPC-SPIOs targeting of M2 macrophages, scavenging of cellular ROS, and activation of magnetic resonance (MR) signals after interactions with cells.
Materials and Method
Synthesis of Uncoated SPIOs, PEGylated SPIOs, IPC-SPIOs, and Mannose-IPC-SPIOs
IPC-SPIOs were synthesized using previously optimized protocols.5,6 Briefly, uncoated SPIOs were synthesized from a mixture of 1 mmol FeCl2 in 50 mL deionized (DI) water (Sigma-Aldrich; St. Louis, MO, USA) and 2 mmol FeCl3 in 50 mL DI water (Sigma; total volume: 100 mL) via the coprecipitation method under N2 atmosphere with vigorous magnetic stirring. The resulting mixture was heated to 80 °C in a silicone oil bath, and 1M of aqueous ammonium hydroxide (NH4OH) (BpDH; Poole, Dorset, UK) was added dropwise and allowed to react for 5 hours to complete the reaction. Uncoated SPIOs were washed and collected through magnetic decantation and centrifugation. SPIOs were coated with PEG by adding PEG 300 kDa (Sigma) into the suspension of uncoated SPIOs (3 mg/mL) in water and stirred using a magnetic stir bar for 24 hours at 1000 rpm. The mixture was purified using magnetic decantation resulting in a final PEG SPIOs concentration of 2 mg/mL. PEG-SPIOs were complexed with poly(gallol) by mixing 0.25 mg/mL aqueous poly(gallol) solution with 2 mg/mL PEG-SPIOs for one hour before adding 1 M of sodium phosphate buffer saline in a 9:9:2 volume ratio of poly(gallol) solution to PEG-SPIOs to sodium phosphate buffer saline. The mixture was left overnight to react before unreacted reagents were removed using magnetic decantation, and the supernatant was collected to be used as untargeted IPC-SPIOs. Mannose-PEG attachment to IPC-SPIOs was achieved by the amine-isothiocyanate reaction. Initially, 6.25 mg/mL and 25 mL of α-D-mannopyranosylphenyl isothiocyanate (Sigma) and O-(2-Aminoethyl)polyethylene glycol hydroxyl (Sigma; molecular weight: 10,000) were separately dissolved in 1 mL DMSO (Sigma). The solutions were mixed before addition of 1 mL sodium bicarbonate buffer (pH 8.5; aqueous). The mixture was allowed to react for 24 hours at room temperature under mild agitation. After 24 hours, the resulting mannose-PEG was added to complexed IPC-SPIOs at a ratio of 1:1 and allowed to mix for 5 minutes. Sodium phosphate buffer was added to the mannose-PEG: IPC-SPIOs mixture and left for 8 hours to induce complexation. After this second complexation, mannose-IPC-SPIOs were washed via centrifugation to remove unreacted reagents, while the supernatant was collected for experiments.
Cell Culture and Differentiation
Human THP-1 (ATCC® TIB71™; Manassas, VA, USA) and RAW264.7 murine cells (ATCC) were cultured in RPMI-1640 (Thermo Fisher Scientific™, Waltham, MA, USA) with L-Glutamine (Sigma), 10% (v/v) fetal bovine serum (FBS) (Seradigm, Radnor, PA, USA), and 1% (v/v) penicillin-streptomycin glutamine (Life Technologies, Burlington, ON, Canada). Both cells were differentiated in T175 tissue culture flasks under standard conditions at 37°C with 5% CO2 and humidity. For cells differentiated into the M1 lineage, cells were treated with 5 ng/mL of granulocyte-macrophage colony-stimulating factor (GM-CSF; Shenandoah Biotechnology, Inc., Warwick, PA, USA) in RPMI for 7 days (media changed every 2–3 days with GM-CSF supplemented RPMI). On the 7th day, cells were treated with 20 ng/mL of interferon gamma (IFN-γ; Shenandoah) for 1–2 hours. Subsequently, cells were also treated with 100 ng/mL lipopolysaccharides (LPS; Sigma) for 24 hours at which time they were ready for use. For cells differentiated into M2 lineages, cells were treated with 25 ng/mL of macrophage colony-stimulating factor (M-CSF; Shenandoah) in RPMI for 7 days (media changed every 2–3 days with M-CSF supplemented RPMI). On the 7th day, cells were treated with interleukin-4 (IL-4; Shenandoah) for 24 hours. On the 8th day, cells were treated with interleukin (IL-13; Shenandoah) for 24 hours at which time they were ready for use.29
Hydrodynamic Size, Zeta Potential, and Polydispersity Index
Dynamic light scattering was used to determine the hydrodynamic size (diameter), zeta potential, and polydispersity index (PDI) of IPC-SPIOs and mannose-IPC-SPIOs using a Zetasizer (Malvern NanoZS, Worcestershire, UK) at room temperature. Samples were suspended either in water or cell media at a nanoparticle concentration of 0.05 mg/mL. All samples were placed in a folded capillary cuvette and inserted into the Zetasizer for testing and analysis.
Analysis of Mannose Incorporation into IPC-SPIOs
First, mannose-PEG attachment was measured using the cyclocondensation of isothiocyanate (ITC) groups present in mannose. This process involved detecting ITC through a chemical reaction of 1,2-benzenodithiol and ITC to form 1,3-benzodithiol-2-thione.30 First, 10 mM of 1,2-benzenedithiol (BDT; Sigma) was dissolved in methanol (total volume 3mL; Sigma). A mixture of 600 μL of BDT, 500 μL of potassium phosphate, and 100 μL of mannose-PEG was vortexed for 1 minute. The mixture was tightly capped in a vial and heated at 65°C for 2 hours. After heating, absorbance was read at 365 nm using a plate reader (Synergy H1, BioTek; Winooski, VT, USA).
In addition to cyclocondensation, to confirm mannose-PEG attachment, Fourier transform-infrared spectroscopy (Compact FT-IR Spectrometer ALPHA II; Bruker; Billerica, MA) was used to investigate the characteristic peaks of mannose and PEG. After mannose-PEG conjugate synthesis, it was placed in a homogeneous solution on the stage of the FT-IR. IR absorptions were collected and analyzed for characteristic peaks of mannose-PEG.
Periodic acid Schiff (PAS) stain was used to quantify the attachment of mannose to IPC-SPIOs. First, 0.6% periodic acid solution was made by adding 0.6 g of periodic acid crystalline powder (Sigma) to 100 mL of ultrafiltered water. Nanoparticles and controls were mixed with 0.6% periodic acid solution at a ratio of 1:5 and allowed to react for 90 minutes. After 90 minutes, Schiff reagent (Sigma) was added to the mixture and allowed to incubate in the dark for an additional 40 minutes. The absorbance of the mixture was read at 550 nm using a plate reader (BioTek), indicating the presence of mannose.
Measurement of Exogenous ROS Scavenging
Xanthine oxidase (0.015 units, XOD, Sigma) in 0.1 M sodium phosphate buffered saline (PBS) was added to 30 mM hypoxanthine (HX, Amresco, Solon, OH, USA) suspended in 50 mM potassium hydroxide (KOH, Research Products International, Mt Prospect, IL, USA) to generate superoxide. Superoxide was added to different concentrations of mannose-IPC-SPIOs and IPC-SPIOs at a ratio of 1:1, and absorbance was measured at 560 nm using a plate reader (BioTek) to detect the nanoparticle change in turbidity.
Histological Staining for Identification of Nanoparticle Attachment to Murine M1/M2 Macrophages
M1 and M2 macrophages were seeded into 96-well polystyrene tissue culture (TC)-treated microplates. Quadruplicate wells were exposed to 150, 100, or 75 μg/mL of IPC-SPIOs or mannose-IPC-SPIOs in cell culture media for 1, 3, or 6 hours. Cell media was aspirated, and cells were rinsed three times with PBS with calcium and magnesium. Controls were cells not exposed to nanoparticles. After the wash procedure, cells were fixed with ice-cold acetone and stained with 2% potassium ferrocyanide II/1 M hydrochloric acid mixture (1:1; Sigma) for 10 min at 37 °C. Cells were counterstained with Nuclear Fast Red solution (0.1%; Sigma) for 1 minute. The cells were imaged with a brightfield microscope (Nikon ECLIPSE Ts2; Tokyo, Japan).
Quantification of Nanoparticle Attachment to Murine M2 Macrophages
M2 macrophages were seeded into 24-well polystyrene TC-treated microplates. Some cells were incubated with 500 μg/mL of mannose (Sigma) for 2 hours to partially block mannose receptors and rinsed twice with PBS with calcium and magnesium. Cells were exposed to 150 μg/mL of mannose-IPC-SPIOs in cell culture media for 1, 3, or 6 hours. Cell media was aspirated, and cells were rinsed twice with PBS supplemented with calcium and magnesium. Control cells were not exposed to nanoparticles. The absorbance of nanoparticles attached to cells was measured at 450 nm using a plate reader (BioTek). Control cells were used to obtain baseline cellular absorbance, and all other data points were compared to this control sample.
Bradford Assay for Quantification of Protein in M1/M2 Macrophages
Both human and mouse M1 and M2 macrophages were seeded into 24-well polystyrene TC-treated microplates. Quadruplicate wells were exposed to 150 μg/mL of PEG-SPIOs, IPC-SPIOs, or mannose-IPC-SPIOs in cell culture media for 6 hours. Cell media was aspirated, and cells were rinsed twice with PBS with calcium and magnesium. Control cells were not exposed to nanoparticles. After the wash procedure, 0.2 mL of ultrafiltered water was added to each well and sonicated for 15 minutes at 4°C. Cells were scraped off the bottom of the well using a pipet tip and transferred into a microcentrifuge tube (VWR Radnor, PA, USA). Cell lysates were further sonicated for 15 minutes at 4°C. After sonication, 5 μL of cell lysate was added to 250 μL of Bradford assay reagent (Sigma) in 96-well clear plates and allowed to incubate for 15 minutes at room temperature. Absorbance was read at 595 nm using a plate reader (BioTek) to detect protein concentration in M1/M2 macrophages.
Inductively Coupled Plasma Optical Emission Spectrometry (ICP-OES) to Quantify Nanoparticles
Both human and murine M1/M2 were seeded into 24-well polystyrene TC-treated microplates. Quadruplicate wells were exposed to 150 μg/mL of PEG-SPIOs, IPC-SPIOs, or mannose-IPC-SPIOs in cell culture media for 6 hours. Cell media was aspirated, and cells were rinsed three times with PBS. Control cells were not exposed to nanoparticles. After the wash procedure, cells were trypsinized (Sigma) and centrifuged at 125 × g for 10 minutes. The cell pellet was digested in 70% nitric acid (Sigma) and reconstituted in 2% nitric acid. Zinc standard was added to the solution as an internal control. Elemental analysis was conducted using Perkin-Elmer Avio 200 (Waltham, MA, USA) in order to detect the concentration of iron taken up by cells.
Cell Viability
Mannose-IPC-SPIOs were diluted to 150, 100, and 75 μg/mL in cell media. Confluent cells were exposed to nanoparticles for 10 hours. After exposure, the cell media was aspirated, and the cells were rinsed three times with PBS. A 110 μL aliquot of diluted cell counting kit-8 (CCK-8, Sigma) reagent (10% CCK-8 in cell media) was added to each well, wells were incubated for 1 hour, and the absorbance was measured at 450 nm to quantify cell viability.
Confocal Imaging
To enable in vitro tracking, nanoparticles were conjugated with FITC to form FITC IPC-SPIOs and FITC mannose-IPC-SPIOs. Briefly, 0.25 mg of FITC-PEG-OH 5000 MW (NANOCS; NY, USA) was added to 2 mg/mL PEG SPIOs solution. The mixture was allowed to stir for 24 hours at 1000 rpm, resulting in FITC PEG SPIOs. Complexed FITC IPC-SPIOs were made by mixing 2 mg/mL FITC PEG SPIOs with 0.25 mg/mL aqueous poly(gallol) solution before adding 1 M of sodium phosphate buffer saline in a 9:9:2 volume ratio of poly(gallol) solution to (FITC)-PEG-SPIOs to sodium phosphate buffer saline. The mixture was left overnight to react before unreacted reagents were removed using magnetic decantation, and the supernatant was collected to be used as untargeted FITC IPC-SPIOs. For targeted nanoparticles, mannose was attached to FITC IPC-SPIOs by adding 2.08 mg/mL of mannose-PEG to FITC IPC-SPIOs at a ratio of 1:1 and allowed to mix for 5 minutes. Sodium phosphate buffer was added to the mannose-PEG: FITC IPC-SPIOs mixture and left for 8 hours to induce complexation. After this second complexation, FITC mannose-IPC-SPIOs were washed via magnetic decantation to remove unreacted reagents, while the supernatant was collected for experiments.
Cells were cultured on a LabTek 8 Chambered Coverglass slide (Thermo Fisher). FITC/non-FITC conjugated IPC-SPIOs (150 μg/mL) were added to wells for 1, 3, and 6 hours of incubation. After incubation, cells were rinsed and fixed with 4% paraformaldehyde at room temperature for 10 minutes. Cells were rinsed again and stained for 10 minutes at 37°C with CellMaskTM Deep Red to stain the plasma membrane (Life Technologies, diluted 1:1000 in PBS). The supernatant was aspirated, cells were rinsed three times with PBS, and the sample was mounted with Prolong Diamond antifade reagent containing 4′,6-diamidino-2-phenylindole (DAPI; Thermo Fisher). Slides were imaged using LSM 880 NOL 3-channel multiphoton confocal laser scanning microscope (Zeiss; Oberkochen, Germany) with the following parameters: ex 633 nm/em 638–755 nm for CellMask deep red and ex 405 nm/em 410–502 nm for DAPI. Images were obtained using a water-immersion 40x objective and oil-immersion 63x objective and Zeiss Efficient Navigation (ZEN) 2.3 software.
T2-Weighted MR Scan
To investigate cellular ROS activation of T2 and T2* MR signals, human and murine M1/M2 macrophages were exposed to 150 μg/mL of PEG-SPIOs, IPC-SPIOs, and mannose-IPC-SPIOs. This was accomplished by culturing and differentiating cells in quadruplicate wells before exposure to nanoparticles in cell culture media for 6 hours. Cell media was aspirated, and cells were rinsed three times with PBS supplemented with calcium and magnesium. Control cells were not exposed to nanoparticles. After the wash procedure, cells were trypsinized, trypsin was deactivated using trypsin neutralizing solution, and cells were added to a custom-built, susceptibility-matched ULTEM 96-well plate and scanned on a 3 T MR scanner using an abdomen coil (Philips 3T Achieva dStream MRI scanner, Amsterdam, Netherlands).
The volume of each sample was maintained at 700 μL in the custom well plate with MR images acquired for a single 5-mm-thick slice with coronal orientation with a field of view of 75 × 75 mm. T2-weighted images were acquired with a multi-spin echo sequence with echo time (TE) = 6.8 ms, echo train length (ETL) = 32, repetition time (TR) = 2000 ms, number of averages (NSA) = 2, and a 90° flip angle resulting in a total scan time of 5.8 minutes. For T2*-weighted images, images were acquired using a multi-gradient echo sequence with 15° flip angle, TE = 1.962 ms, ETL = 16, pixels, TR = 200 ms and NSA = 2, resulting in a total scan time of 3.3 minutes. After acquiring MR images, regions of interest (ROI) were drawn on samples on the 96-well plate. The pixel intensity from the ROI was averaged as a function of echo time (TE) and fit into T2 and T2* equations using MATLAB® (MathWorks, Natick, MA, USA).
Results and Discussion
Optimization of Targeting Biomolecule
The conjugation of mannose to amine PEG occurred at the ITC component of mannose via amine-isothiocyanate coupling (Figure 1A). The amine-isothiocyanate reaction resulted in the formation of a thiourea, which links mannose to amine-PEG.31 Cyclocondensation of thiourea using 1,2-benzenedithiol to produce 1,3-benzodithiol-2-thione resulted in a higher absorbance value compared to cyclocondensation of isothiocyanate. The absorbance values of mannose-PEG were twice as high as mannose containing isothiocyanate (Figure 1B). As expected, sodium bicarbonate, PEG, and mannose-PEG without 1,2-benzenodithiol did not produce any appreciable absorbance values. This study provided evidence of mannose-PEG attachment, which was further confirmed by FT-IR. FT-IR detected peaks for the C-O bonds and C-OH band at 1000 cm−1–1075 cm−1 and 1059 cm−1, ether rings, hydroxyl, and C-N bonds in PEG at 850 cm−1–775 cm−1, 3400 cm−1–3200 cm−1, and 1320 cm−1–1200 cm−1 respectively. The formation of thiourea was also confirmed with characteristic peaks for amine, C-N, and C=S bonds at 3362 cm−1, 1093 cm−1, and 775 cm−1 (Figure 1C). These peaks indicated mannose-PEG attachment, thereby supporting cyclocondensation data that suggested successful mannose-PEG attachment.
Nanoparticle Characterization
Full characterization of IPC-SPIOs was published previously.6,32 Mannose attachment to IPC-SPIOs was investigated using carbohydrate staining of nanoparticles. Mannose showed a concentration-dependent attachment to IPC-SPIOs. A linear relationship was exhibited between 75, 150, and 300 μg/mL of IPC-SPIOs conjugated to 2.08 mg/mL of mannose with an R2 linear fit value of 0.997 (Figure 2A). Further, a linear relationship was also evident between 2.08, 1.04, and 0.52 mg/mL of mannose when the concentration of IPC-SPIOs was held constant at 300 μg/mL (Figure 2B). These results established mannose-PEG attachment to IPC-SPIOs via complexation and confirmed that mannose attaches to IPC-SPIOs in a concentration-dependent manner.
After incorporating mannose, the hydrodynamic size, zeta potential, and polydispersity index (PDI) of 0.05 mg/mL of IPC-SPIOs and mannose-IPC-SPIOs were investigated using dynamic light scattering (DLS) in water or cell media. The incorporation of mannose into IPC-SPIOs triggered an increase in the hydrodynamic size from 118 nm to 131.2 nm (Table 1). The zeta potential also moved closer to neutral after surface modification by mannose. The hydrodynamic size of IPC-SPIOs and mannose-IPC-SPIOs in cell media were much higher compared to nanoparticles in water, likely due to the adsorption of serum proteins. Nanoparticles in water had a higher negative zeta potential and lower PDI compared to nanoparticles in cell media. The difference in zeta potential and PDI could also be attributed to serum proteins interfering with nanoparticle aggregation. While the particles in media are likely too large for intravenous administration, their size is appropriate for their planned use in intra-articular injections to assess joint inflammation.
 |
Table 1 Hydrodynamic Sizes, Zeta Potential, and Polydispersity Indexes of Complexed IPC-SPIOs and Mannose-IPC-SPIOs in Water and Media Measured Using Dynamic Light Scattering (DLS n = 4) |
Exogenous ROS Scavenging by Mannose-IPC-SPIOs
Attachment of mannose to IPC-SPIOs altered the surface of the nanoparticle significantly, which was also hypothesized to change ROS scavenging behavior. Exogenous superoxide was used to investigate ROS scavenging by mannose-IPC-SPIOs. Previously, we established that among species of ROS, IPC-SPIOs had the greatest scavenging effect on superoxide, with the highest concentration of IPC-SPIOs decomplexing by as much as 95% after interaction with available superoxide.5 Surface modification by attachment of mannose resulted in a slight reduction of mannose-IPC-SPIOs decomplexation, which appeared to be 80% after 6 hours compared to the 95% detected in IPC-SPIOs (Figure 3A). However, at lower concentrations, mannose-IPC-SPIOs decomplexation was similar to untargeted IPC-SPIOs. The rate of mannose-IPC-SPIOs scavenging appeared to be concentration-dependent (Figure 3B), a trend similar to superoxide scavenging by IPC-SPIOs. These trends provide evidence of similarities between IPC-SPIOs and mannose-IPC-SPIOs ROS scavenging behavior.
Nanoparticle Uptake by Murine Macrophages
After polarization into M1 and M2 lineages, Prussian blue histological staining was used to detect iron as a measure of IPC-SPIO nanoparticle attachment to both macrophage lineages. Cells in vitro were stained in a manner similar to the widely used Perl Prussian blue staining procedure of tissues to detect iron content. M1 and M2 macrophages treated with IPC-SPIOs and mannose-IPC-SPIOs were a deep blue color after staining (Figure S1 A-B). For M2 macrophages, the intensity of blue color was time-dependent, with more blue color detected as nanoparticle exposure time increased (Figure S1B). Nanoparticle attachment to M1 macrophages did not show any discernable trend.
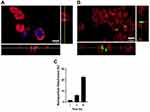
After establishing nanoparticle attachment to cells, intracellular uptake was investigated using confocal microscopy. This study was conducted on M1 and M2 macrophages exposed to 150 μg/mL of FITC-conjugated IPC-SPIOs for 6 hours. Cells were stained with the nuclear stain DAPI and cell membrane stain CellMaskTM Red, and samples were imaged using confocal microscopy to determine the uptake of nanoparticles. Intracellular localization of the fluorescent signal from nanoparticles was detected within the cell membrane of both M1 and M2 macrophages, although uptake differed significantly between samples (Figure 4A–B and S2A–S2B). Nanoparticles were confirmed inside cells via orthogonal projections of cells, and M2 macrophages exposed to mannose-IPC-SPIOs had the most intracellular FITC signal amongst the experimental conditions (Figure 4B).
To quantify uptake, some M2 macrophages were pretreated with 500 µg/mL of mannose to inhibit mannose receptors, while the mannose receptors for the remaining M2 macrophages were not inhibited. Mannose receptor-mediated mannose-IPC-SPIOs uptake was confirmed by measuring the absorbance of nanoparticles on cells. The attachment of mannose-IPC-SPIOs to M2 macrophages pre-treated with mannose was significantly restricted compared to M2 macrophages with free mannose receptors. Within 1–3 hours of mannose-IPC-SPIOs treatment, M2 macrophages with free mannose receptors experienced over 6.1% more localization of mannose-IPC-SPIOs compared to M2 macrophages pre-treated with mannose (Figure 4C). As nanoparticle exposure time was extended to 6 hours, nanoparticle localization to M2 macrophages with free mannose receptors was over 22% higher than M2 macrophages pre-treated with mannose (Figure 4C). These results provided strong evidence of time-dependent receptor-mediated endocytosis of mannose-IPC-SPIOs.
M1 macrophages showed uptake of IPC-SPIOs and mannose-IPC-SPIOs. Uptake was visibly lower compared to levels observed in M2 macrophages (Figure S2A and S2B). This confirmed histological studies that suggested both IPC-SPIOs and mannose-IPC-SPIOs attached to M1 and M2 macrophages; however, nanoparticle attachment is more pronounced in M2 macrophages. The uptake of IPC-SPIOs by M1 and M2 macrophages was expected to be via clathrin-mediated endocytosis because of the size of the particle. Foroozandeh et al reported that nanoparticles with a maximum size of 150 nm are usually taken up via clathrin-mediated endocytosis.33 This pathway is triggered by an interaction between agonist and cell receptor leading to the formation of endocytic clathrin-coated vesicles of sizes 70–150 nm. After internalization, the vesicle loses its clathrin coating and fuses with other vesicles, triggering a cascade of events that result in the delivery to a lysosome or intracellular site.
The uptake of mannose-IPC-SPIOs by M1 macrophages may rely on phagocytosis because of the larger hydrodynamic size of the particles. In contrast, M2 macrophages likely internalize mannose-IPC-SPIOs via both phagocytosis and receptor-mediated endocytosis because of the presence of mannose receptors.34–36 In receptor-mediated endocytosis, the binding ligand deposits nanoparticles to a specific receptor causing a conformational change in the plasma membrane.30 Nanoparticles are internalized by the invagination of the plasma membrane, which generates early endosomes.37 For mannose receptor-mediated endocytosis, the mannose receptor contains eight extracellular C-type lectin-like domains (CTLDs).38 Mannose ligands cluster multiple glycosides together, thereby enabling multivalent interactions with multidomain lectin receptors (mannose receptors), significantly increasing overall avidity.31 The clustering of mannose-IPC-SPIOs on mannose multidomain lectin receptors would trigger multivalent interactions with mannose, initiating significant uptake of nanoparticles.

Mf = Nanoparticle on cells with free mannose receptor
Mb = Nanoparticles on cells with blocked mannose receptor.
Nanoparticle Uptake as Measured by ICP-OES for Human and Murine Macrophages
The amount of nanoparticle uptake was investigated with inductively coupled plasma optical emission spectroscopy (ICP-OES). For this study, cells treated with 150 ug/mL of IPC-SPIOs and mannose-IPC-SPIOs were digested to extract iron content. Zinc was used as an internal control for ICP-OES, while cells in fresh media were used as a control for nanoparticle uptake. The Bradford assay was used to determine the concentration of proteins per well in the tissue culture plate, which allowed for normalization of data to the amount of protein in cellular samples instead of counted cells.
The highest uptake was detected in M2 mouse macrophages treated with mannose-IPC-SPIOs. Iron detected in M2 mouse macrophages was 21.4 ± 3.94 parts per million (ppm) when exposed to mannose-IPC-SPIOs compared to IPC-SPIOs at 7.041 ± 2.24 ppm (Figure 5A). M1 mouse macrophages had lower nanoparticle uptake, with concentrations of IPC-SPIOs and mannose-IPC-SPIOs reported as 6.02 ± 3.07 ppm and 4.15 ± 2.9 ppm, respectively (Figure S3A). Human macrophages had a lower uptake rate across all experimental conditions than mouse macrophages. Nonetheless, like mouse macrophages, the highest iron content was detected in M2 human macrophages exposed to mannose-IPC-SPIOs. The concentration of iron from mannose-IPC-SPIOs detected in M2 human macrophages was 623 ± 12.479 parts per billion (ppb) compared to 449 ± 53.670 ppb for IPC-SPIOs (Figure 5B). In M1 human macrophages, iron concentrations from IPC-SPIOs and mannose-IPC-SPIOs were also lower than in M2 human macrophages, with concentrations of 287 ± 12 ppb and 375 ± 12.163 ppb, respectively (Figure S3B).
The disparity between nanoparticle uptake between human and mouse macrophages can be attributed to inherent differences between both cell types. In this study, RAW264.7 was differentiated into mouse M1 and M2 macrophages, while THP-1 was differentiated into human M1 and M2 macrophages. Martinez-Marin et al showed that endocytic activity in RAW264.7 is much higher than THP-1.29 In that study, unstimulated RAW264.7 had over ten times more endocytic activity than unstimulated THP-1. RAW264.7 is primarily designed for endocytic assays and has been optimized significantly to enhance uptake capability. These factors contribute to the differences between uptake detected in the two cell lines used in these studies.
Cell Viability
Cellular viability was investigated to determine the toxicity of mannose-IPC-SPIOs on M2 macrophages. Several concentrations of mannose-IPC-SPIOs were exposed to M2 mouse and human macrophages for 10 hours. The colorimetric CCK-8 assay was used to investigate the effects of nanoparticles on cells. Cells only, which serve as a control, are cells in fresh media without particle exposure. Compared to controls, M2 macrophages showed no statistically significant reduction in viability after exposure to any of the particles tested at 150 μg/mL, 100 μg/mL, or 75 μg/mL (Figure 6).
Cellular Activation of MR Signals
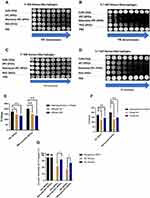
Activation of T2 MR signal by cellular ROS was investigated with 3 T MR scanner. First, IPC-SPIOs, mannose-IPC-SPIOs, decomplexed IPC-SPIOs, and decomplexed mannose-IPC-SPIOs were scanned to confirm the activation of complexed nanoparticles after exposure to ROS (Figure S5). Thereafter, M1 and M2 macrophages were exposed to IPC-SPIOs or mannose-IPC-SPIOs. After 6 hours, T2 MR scans were obtained to investigate cellular ROS-mediated contrast activation by targeted and untargeted IPC-SPIOs. MR contrast was detected in cells treated with nanoparticles compared to cells alone (Figure 7A and C). The greatest change in MR contrast was detected in M2 macrophages treated with mannose-IPC-SPIOs compared to IPC-SPIOs (Figure 7A and C). MR contrast activation was also detected in M1 macrophages treated with IPC-SPIOs and mannose-IPC-SPIOs, although contrast was much weaker compared to signals detected in M2 macrophages (Figure S4A–S4D). T2* MR signals also showed nanoparticle-induced contrast in cells. M2 macrophages treated with mannose-IPC-SPIOs also induced the highest T2* contrast compared to other experimental conditions (Figure 7B and D).
The significant MR contrast detected in M2 macrophages treated with mannose-IPC-SPIOs was attributed to mannose receptor-mediated nanoparticle uptake. The mannose receptor-mediated nanoparticle uptake and ROS scavenging are validated, given the high MR contrast detected in M2 macrophages exposed to mannose-IPC-SPIOs. ROS-induced activatable MR contrast was achieved through interactions between the poly(gallol) coating on the nanoparticle and ROS, as we have previously published.5,6 The addition of poly(gallol) increased T2 values significantly, thereby shielding the contrasting ability of SPIOs. Reaction with cellular ROS can reverse the shielding process, although not completely, by disrupting the hydrogen bonds between PEG and poly(gallol). Disruptions to the hydrogen bond reduce the T2 value and move those values closer to the original T2 value of PEG-SPIOs. This process forms the basis of ROS scavenging by IPC-SPIOs.
The T2 values of cells treated with nanoparticles reflected the disparity in cell-mediated nanoparticle activation between M1 and M2 cells. M2 mouse macrophages treated with mannose-IPC-SPIOs had the highest loss of MR signal, with T2 values ranging from 201 ms to 137 ms pre- to post-cellular ROS exposure (Figure 7E). In contrast, M2 mouse macrophages treated with IPC-SPIOs had pre- to post-cellular ROS exposure T2 values of 174 ms to 136 ms (Figure 7E). The percent of MR signal activation was 22% for IPC-SPIOs and 32% for mannose-IPC-SPIOs (Figure 7G). For human M2 macrophages, the nanoparticle activation trends were similar. T2 values for M2 human macrophages treated with mannose-IPC-SPIOs ranged from 201 ms to 147 ms pre- to post-cellular ROS exposure, while T2 values for IPC-SPIOs ranged from 174 ms to 143 ms pre- to post-cellular ROS exposure (Figure 7F). The percent activation for IPC-SPIOs and mannose-IPC-SPIOs were 18% and 27%, respectively (Figure 7G). T1 MR contrast is not reported here because Yoo et al previously showed that IPC-SPIOs do not appreciably activate T1 contrast.6 In that study, the T1 relaxivity (r1) of complexed IPC-SPIOs and decomplexed IPC-SPIOs ranged from 0.7 mM−1·s−1 to 1.14 mM−1·s−1 in superoxide compared to T2 relaxivity that ranged from 7 mM−1·s−1 to 70 mM−1·s−1 respectively.6 It was concluded in that study that complexation of poly(gallol) on SPIOs affected T1 contrasting; however, decomplexation and activation of T1 signal was very poor compared to T2.
Conclusion
This study demonstrated surface modification of IPC-SPIOs to incorporate mannose as a targeting biomolecule to improve localization, uptake, and activation of MR contrast agents. The functionalized nanoparticle targeted mannose receptors efficiently and showed time-dependent uptake of nanoparticles. Magnetic resonance was used to confirm receptor-mediated ROS activation of T2 signals, and M2 macrophages exposed to mannose-IPC-SPIOs experienced the greatest MR contrast compared to other experimental conditions. These results show that mannose-IPC-SPIOs specifically target M2 macrophages and scavenge cellular ROS to activate the T2 MR signal.
Acknowledgments
Financial support for this work was provided by the NIH (1R21EB017504-01) and Clifford D Clark Diversity Fellowship at Binghamton University. Equipment usage was supported by the Binghamton University Analytical and Diagnostics Laboratory and University of Vermont-MRI Center for Biomedical Imaging. The authors thank Sebastian Freeman for support with confocal imaging and image analysis. The authors disclose that information presented in this research article has already been published in Chukwuazam Nwasike’s dissertation (Nwasike, C. A. (2020). Targeted activatable magnetic resonance (MR) contrast agents sensitive to inflammation induced oxidative stress (Order No. 28259582) (2512722915). Retrieved from https://www.proquest.com/dissertations-theses/targeted-activatable-magnetic-resonance-mr/docview/2512722915/se-2).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Fujiwara N, Kobayashi K. Macrophages in inflammation. Curr Drug Targets. 2005;4(3):281–286. doi:10.2174/1568010054022024
2. Krzyszczyk P, Schloss R, Palmer A, Berthiaume F. The role of macrophages in acute and chronic wound healing and interventions to promote pro-wound healing phenotypes. Front Physiol. 2018;9:419. doi:10.3389/fphys.2018.00419
3. Orecchioni M, Ghosheh Y, Pramod AB, Ley K. Macrophage polarization: different gene signatures in M1 (LPS+) vs. classically and M2 (LPS–) vs. alternatively activated macrophages. Front Immunol. 2019;10:1084. doi:10.3389/fimmu.2019.01084
4. Nwasike C, Purr E, Yoo E, Nagi JS, Doiron AL. Activatable nanoparticles: recent advances in redox-sensitive magnetic resonance contrast agent candidates capable of detecting inflammation. Pharmaceuticals. 2021;14(1):69. doi:10.3390/ph14010069
5. Nwasike C, Yoo E, Purr E, Doiron AL. Activatable superparamagnetic iron oxide nanoparticles scavenge reactive oxygen species in macrophages and endothelial cells. RSC Adv. 2020;10(68):41305–41314. doi:10.1039/D0RA06683D
6. Yoo E, Cheng HA, Nardacci LE, et al. Activatable interpolymer complex-superparamagnetic iron oxide nanoparticles as magnetic resonance contrast agents sensitive to oxidative stress. Colloids Surf B Biointerfaces. 2017;158:578–588. doi:10.1016/j.colsurfb.2017.07.025
7. Friedman D, Claypool S, Liu R. The smart targeting of nanoparticles. Curr Pharm Des. 2013;19(35):6315–6329. doi:10.2174/13816128113199990375
8. Rosenblum D, Joshi N, Tao W, Karp JM, Peer D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat Commun. 2018;9(1):1–12. doi:10.1038/s41467-018-03705-y
9. Schmid D, Park CG, Hartl CA, et al. T cell-targeting nanoparticles focus delivery of immunotherapy to improve antitumor immunity. Nat Commun. 2017;8(1):1–12. doi:10.1038/s41467-017-01830-8
10. Yougbaré S, Mutalik C, Okoro G, et al. Emerging trends in nanomaterials for antibacterial applications. Int J Nanomedicine. 2021;16:5831–5867. doi:10.2147/ijn.S328767
11. Kuo J-C, Tan S-H, Hsiao Y-C, et al. Unveiling the antibacterial mechanism of gold nanoclusters via in situ transmission electron microscopy. ACS Sustain Chem Eng. 2022;10(1):464–471. doi:10.1021/acssuschemeng.1c06714
12. Chang T-K, Cheng T-M, Chu H-L, et al. Metabolic mechanism investigation of antibacterial active cysteine-conjugated gold nanoclusters in Escherichia coli. ACS Sustain Chem Eng. 2019;7(18):15479–15486. doi:10.1021/acssuschemeng.9b03048
13. Sharma V, Ichikawa M, Freeze HH. Mannose metabolism: more than meets the eye. Biochem Biophys Res Commun. 2014;453(2):220–228. doi:10.1016/j.bbrc.2014.06.021
14. Jaynes JM, Sable R, Ronzetti M, et al. Mannose receptor (CD206) activation in tumor-associated macrophages enhances adaptive and innate antitumor immune responses. Sci Transl Med. 2020;12:530. doi:10.1126/scitranslmed.aax6337
15. Diebold SS, Plank C, Cotten M, Wagner E, Zenke M. Mannose receptor-mediated gene delivery into antigen presenting dendritic cells. Somat Cell Mol Genet. 2002;27(1):65–74. doi:10.1023/A:1022975705406
16. Tsuchiya K, Suzuki Y, Yoshimura K, et al. Macrophage mannose receptor CD206 predicts prognosis in community-acquired pneumonia. Sci Rep. 2019;9(1):1–10. doi:10.1038/s41598-019-55289-2
17. Apostolopoulos V, McKenzie I. Role of the mannose receptor in the immune response. Curr Mol Med. 2001;1(4):469–474. doi:10.2174/1566524013363645
18. Italiani P, Boraschi D. From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front Immunol. 2014;5:514. doi:10.3389/fimmu.2014.00514
19. Liu J, Geng X, Hou J, Wu G. New insights into M1/M2 macrophages: key modulators in cancer progression. Cancer Cell Int. 2021;21(1):389. doi:10.1186/s12935-021-02089-2
20. Mills CD. M1 and M2 macrophages: oracles of health and disease. Crit Rev Immunol. 2012;32(6):463–488. doi:10.1615/critrevimmunol.v32.i6.10
21. Yunna C, Mengru H, Lei W, Weidong C. Macrophage M1/M2 polarization. Eur J Pharmacol. 2020;877:173090. doi:10.1016/j.ejphar.2020.173090
22. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–969. doi:10.1038/nri2448
23. Wang L, Zhang S, Wu HJ, Rong X, Guo J. M2b macrophage polarization and its roles in diseases. J Leukoc Biol. 2019;106(2):345–358. doi:10.1002/JLB.3RU1018-378RR
24. Asai A, Tsuchimoto Y, Ohama H, et al. Host antitumor resistance improved by the macrophage polarization in a chimera model of patients with HCC. Oncoimmunology. 2017;6(4):e1299301. doi:10.1080/2162402X.2017.1299301
25. Ito I, Bhopale KK, Nishiguchi T, et al. The polarization of M2b monocytes in cultures of burn patient peripheral CD14+ cells treated with a selected human CCL1 antisense oligodeoxynucleotide. Nucleic Acid Ther. 2016;26(5):269–276. doi:10.1089/nat.2016.0617
26. Nishiguchi T, Ito I, Lee JO, Suzuki S, Suzuki F, Kobayashi M. Macrophage polarization and MRSA infection in burned mice. Immunol Cell Biol. 2017;95(2):198–206. doi:10.1038/icb.2016.84
27. Gensel JC, Zhang B. Macrophage activation and its role in repair and pathology after spinal cord injury. Brain Res. 2015;1619:1–11. doi:10.1016/j.brainres.2014.12.045
28. Yue Y, Yang X, Feng K, et al. M2b macrophages reduce early reperfusion injury after myocardial ischemia in mice: a predominant role of inhibiting apoptosis via A20. Int J Cardiol. 2017;245:228–235. doi:10.1016/j.ijcard.2017.07.085
29. Zhang M, Hutter G, Kahn SA, et al. Anti-CD47 treatment stimulates phagocytosis of glioblastoma by M1 and M2 polarized macrophages and promotes M1 polarized macrophages in vivo. PLoS One. 2016;11(4):e0153550. doi:10.1371/journal.pone.0153550
30. Zhang Y. The 1, 2-benzenedithiole-based cyclocondensation assay: a valuable tool for the measurement of chemopreventive isothiocyanates. Crit Rev Food Sci Nutr. 2012;52(6):525–532. doi:10.1080/10408398.2010.503288
31. Lindhorst TK, Kieburg C. Glycocoating of Oligovalent amines: synthesis of thiourea‐bridged cluster glycosides from glycosyl isothiocyanates. Angewandte Chemie Int Edition. 1996;35(17):1953–1956. doi:10.1002/anie.199619531
32. Yoo E, Liu Y, Nwasike CA, et al. Surface characterization of nanoparticles using near-field light scattering. Beilstein J Nanotechnol. 2018;9:1228–1238. doi:10.3762/bjnano.9.114
33. Foroozandeh P, Aziz AA. Insight into cellular uptake and intracellular trafficking of nanoparticles. Nanoscale Res Lett. 2018;13(1):1–12. doi:10.1186/s11671-018-2728-6
34. Gustafson HH, Holt-Casper D, Grainger DW, Ghandehari H. Nanoparticle uptake: the phagocyte problem. Nano Today. 2015;10(4):487–510. doi:10.1016/j.nantod.2015.06.006
35. Jahagirdar P, Lokhande AS, Dandekar P, Devarajan PV. Mannose receptor and targeting strategies. In: Targeted Intracellular Drug Delivery by Receptor Mediated Endocytosis. Springer; 2019:433–456.
36. Martinez-Marin D, Jarvis C, Nelius T, Filleur S. Assessment of phagocytic activity in live macrophages-tumor cells co-cultures by Confocal and Nomarski Microscopy. Biol Methods Protocols. 2017;2(1):bpx002. doi:10.1093/biomethods/bpx002
37. Manzanares D, Ceña V. Endocytosis: the nanoparticle and submicron nanocompounds gateway into the cell. Pharmaceutics. 2020;12(4):371. doi:10.3390/pharmaceutics12040371
38. Song E-H, Manganiello MJ, Chow Y-H, et al. In vivo targeting of alveolar macrophages via RAFT-based glycopolymers. Biomaterials. 2012;33(28):6889–6897. doi:10.1016/j.biomaterials.2012.06.025