Liquid Biopsy Test: Protocol and Steps
Want to listen to this article for FREE?
Complete the form below to unlock access to ALL audio articles.
In this article, explore some of the advanced molecular techniques that are most used for detecting the main biomarkers involved in liquid biopsy.
How does a liquid biopsy work?
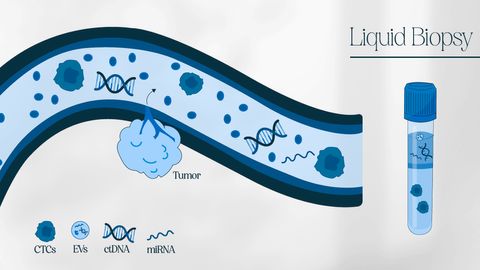
A liquid biopsy consists of the isolation and characterization of tumor-derived materials – such as intact cancer cells or nucleic acids – in a sample of body fluid collected from a cancer patient to gain information about their disease.1
Since most tumors are in contact with the bloodstream, liquid biopsies often involve blood sample analysis – but other sources of body fluids include urine, stool, saliva, pleural fluid, cerebral spinal fluid and peritoneal fluid or washings.2
Liquid biopsy targets
The list of potential tumor-derived materials that can be analyzed in a liquid biopsy includes:
- Circulating tumor cells (CTCs)3 – intact cancer cells shed by both primary and metastatic tumors.
- Circulating cell-free DNA (cfDNA)4 and circulating tumor DNA (ctDNA)–fragments of DNA released from cells (cfDNA), and the tumor-derived DNA fraction (ctDNA).
- Extracellular vesicles (EVs)5– small membrane-bound vesicles, including micro-vesicles and exosomes, which can carry cargo – such as nucleic acids and proteins – during tumor metastasis.
- Circulating cell-free RNA (cfRNA) – cancer-related messenger RNAs or non-coding RNAs, including microRNAs (miRNAs).
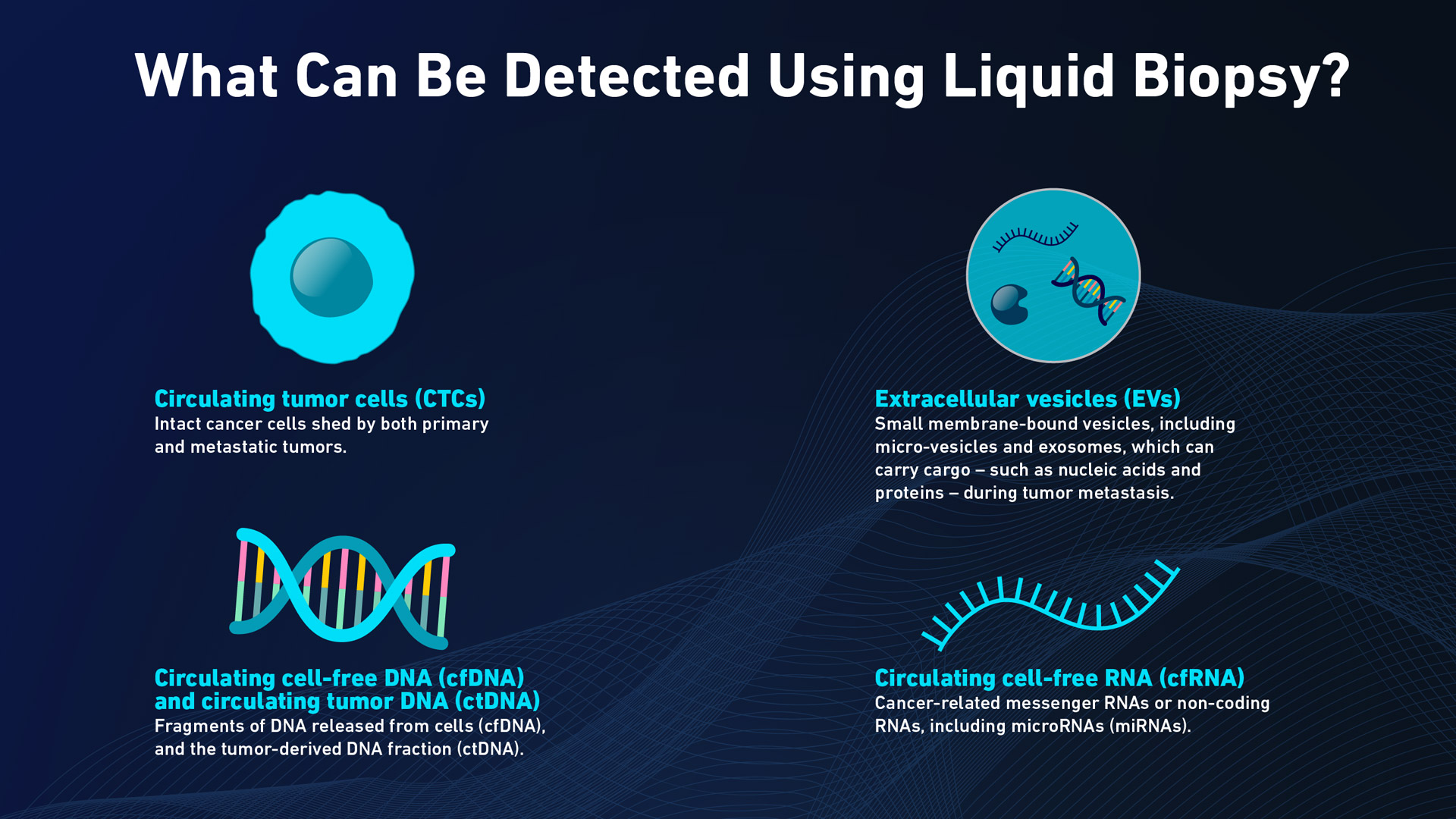
The tumor-derived materials that liquid biopsy can detect. Credit: Technology Networks
Liquid biopsy protocol and steps
Liquid biopsy protocols are focused on the rapid detection of clinically relevant information with high sensitivity, selectivity and specificity while remaining minimally invasive.
The techniques selected for a liquid biopsy will depend on the nature of the target. For instance, it may involve the isolation, enumeration and characterization of CTCs – or the isolation and molecular analysis of other tumor-derived materials.
CTCs
1. Isolation
Over the last decade, several technologies have emerged that aim to isolate and separate CTCs from the massive pool of circulating blood cells – and then count and characterize them.3 Each of these approaches exploits a key difference that discriminates CTCs from normal blood cells (such as differential gene/protein expression, morphology, volume or biophysical properties) to enable the sensitive selection and capture of these very rare cells. They include:
Immune-based detection methods – these are the most widely used approaches and use antibodies to selectively bind cell-surface antigens that discriminate CTCs from blood cells. These antibodies may be conjugated to magnetic nanoparticles7 or immobilized on the walls of microfluidic chips8 to isolate CTCs. A new generation of CTC antigen-based dual modality platforms combines immunomagnetic beads with microfluidics.
- Size-dependent isolation techniques – including filtration, microfluidics, centrifugation and inertial focusing.
- Direct imaging – imaging-based detection methods use specific fluorescent tags to identify and count CTCs in blood. Each of these technologies is unique in its sample preparation, detection algorithms and fluorophores.
- Dielectrophoresis – which relies on particles with different polarization that move differently under a non-uniform electric field.
2. Enumeration
The methods used to count the number of CTCs detected in a sample include impedance10, high-throughput imaging11, flow cytometry12 and artificial intelligence.
3. Characterization
Techniques used to characterize CTCs include immunostaining, fluorescent in situ hybridization (FISH), sequencing, quantitative reverse-transcription polymerase chain reaction (qRT-PCR), expression analysis and cell culture.3
EVs
The clinical application of EVs detected by liquid biopsy has so far been limited by the lack of simple, efficient procedures to obtain EVs with high purity.13 Some of the major strategies used to isolate these extracellular vesicles include ultracentrifugation, polymer precipitation, ultrafiltration, size-exclusion chromatography, affinity isolation and microfluidics-based techniques.13 Each of these approaches has its advantages and disadvantages.
cfDNA/ctDNA
Several methods have been used to quantify and detect tumor mutations in cfDNA/ctDNA14 – including droplet digital PCR15, beads emulsion, amplification and magnetics (BEAMing)16, next-generation sequencing-based approaches17 (whole exome or whole genome sequencing), cancer personalized profiling by deep sequencing (CAPP-seq)16 and tagged-amplicon deep sequencing (TAm-Seq).18 The profiling of epigenetic features of ctDNA, such as methylation signatures19, may provide information about the tissue of origin.
cfRNA
Profiling circulating cell-free (cfRNA) offers unique opportunities to detect cancer, predict the tumor of origin, and determine the molecular subtype.20 But technical and biological variability, coupled with a lack of standardized protocols, have so far hindered the clinical development of RNA-based liquid biopsies.21
References
- Lone SN, Nisar S, Masoodi T, et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer. 2022;21(1):79. doi:10.1186/s12943-022-01543-7
- Satyal U, Srivastava A and Abbosh P. Urine Biopsy-Liquid Gold for Molecular Detection and Surveillance of Bladder Cancer. Front Oncol. 2019;19(9):1266. doi: 10.3389/fonc.2019.01266.
- Habli Z, AlChamaa W, Saab R, Kadara H, Khraiche ML. Circulating Tumor Cell Detection Technologies and Clinical Utility: Challenges and Opportunities. Cancers (Basel). 2020;12(7):1930. doi:10.3390/cancers12071930
- Yan Y, Guo Q, Wang F, et al. Cell-Free DNA: Hope and Potential Application in Cancer. Front Cell Dev Biol. 2021;9:639233. doi:10.3389/fcell.2021.639233
- Qiao F, Pan P, Yan J, et al. Role of tumor‑derived extracellular vesicles in cancer progression and their clinical applications (Review). Int J Oncol. 2019;54(5):1525-1533. doi:10.3892/ijo.2019.4745
- Roskams-Hieter B, Kim HJ, Anur P, et al. Plasma cell-free RNA profiling distinguishes cancers from pre-malignant conditions in solid and hematologic malignancies. npj Precis Onc. 2022;6(1):1-11. doi:10.1038/s41698-022-00270-y
- Su D, Wu K, Saha R, Liu J, Wang JP. Magnetic nanotechnologies for early cancer diagnostics with liquid biopsies: a review. J Cancer Metastasis Treat. 2020;6:19. doi:10.20517/2394-4722.2020.48
- Zou D, Cui D. Advances in isolation and detection of circulating tumor cells based on microfluidics. Cancer Biol Med. 2018 15(4):335-353. doi: 10.20892/j.issn.2095-3941.2018.0256.
- Russo GI, Musso N, Romano A, et al. The Role of Dielectrophoresis for Cancer Diagnosis and Prognosis. Cancers (Basel). 2021;14(1):198. doi:10.3390/cancers14010198
- Ghassemi P, Ren X, Foster BM, Kerr BA, Agah M. Post-enrichment circulating tumor cell detection and enumeration via deformability impedance cytometry. Biosens. Bioelectron. 2020;150:111868. doi:10.1016/j.bios.2019.111868
- Keomanee-Dizon K, Shishido S and Kuhn P. Circulating Tumor Cells: High-Throughput Imaging of CTCs and Bioinformatic Analysis. Recent Results Cancer Res. 2020;215:89-104. doi: 10.1007/978-3-030-26439-0_5.
- Lopresti A, Malergue F, Bertucci F, et al. Sensitive and easy screening for circulating tumor cells by flow cytometry. JCI Insight. 4(14):e128180. doi:10.1172/jci.insight.128180
- Liu J, Chen Y, Pei F, et al. Extracellular Vesicles in Liquid Biopsies: Potential for Disease Diagnosis. BioMed Research International. 2021;2021:e6611244. doi:10.1155/2021/6611244
- Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. Journal of Hematology & Oncology. 2022;15(1):131. doi:10.1186/s13045-022-01351-y
- Kaneko A, Kanemaru H, Kajihara I, et al. Liquid biopsy-based analysis by ddPCR and CAPP-Seq in melanoma patients. J Dermatol Sci. 2021;102(3):158-166. doi:10.1016/j.jdermsci.2021.04.006
- García-Foncillas J, Alba E, Aranda E, et al. Incorporating BEAMing technology as a liquid biopsy into clinical practice for the management of colorectal cancer patients: an expert taskforce review. Ann Oncol. 2017;28(12):2943-2949. doi:10.1093/annonc/mdx501
- Lin C, Liu X, Zheng B, Ke R, Tzeng CM. Liquid Biopsy, ctDNA Diagnosis through NGS. Life (Basel). 2021;11(9):890. doi:10.3390/life11090890
- Gale D, Lawson ARJ, Howarth K, et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS One. 2018;13(3):e0194630. doi:10.1371/journal.pone.0194630
- Li P, Liu S, Du L, Mohseni G, Zhang Y, Wang C. Liquid biopsies based on DNA methylation as biomarkers for the detection and prognosis of lung cancer. Clin. Epigenetics. 2022;14(1):118. doi:10.1186/s13148-022-01337-0
- Larson MH, Pan W, Kim HJ, et al. A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection. Nat Commun. 2021;12(1):2357. doi:10.1038/s41467-021-22444-1
- Cabús L, Lagarde J, Curado J, Lizano E, Pérez-Boza J. Current challenges and best practices for cell-free long RNA biomarker discovery. Biomarker Research. 2022;10(1):62. doi:10.1186/s40364-022-00409-w



